Abstract
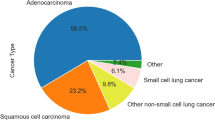
Lung cancer is the leading cause of cancer death worldwide. Survival is largely dependent on the stage of diagnosis: the localized disease has a 5-year survival greater than 55%, whereas, for spread tumors, this rate is only 4%. Therefore, the early detection of lung cancer is key for improving prognosis. In this study, we present an innovative, non-invasive, cancer detection approach based on measurements of the metabolic activity profiles of immune system cells. For each Liquid ImmunoBiopsy test, a 384 multi-well plate is loaded with freshly separated PBMCs, and each well contains 1 of the 16 selected stimulants in several increasing concentrations. The extracellular acidity is measured in both air-open and hermetically-sealed states, using a commercial fluorescence plate reader, for approximately 1.5 h. Both states enable the measurement of real-time accumulation of ‘soluble’ versus ‘volatile’ metabolic products, thereby differentiating between oxidative phosphorylation and aerobic glycolysis. The metabolic activity profiles are analyzed for cancer diagnosis by machine-learning tools. We present a diagnostic accuracy study, using a multivariable prediction model to differentiate between lung cancer and control blood samples. The model was developed and tested using a cohort of 200 subjects (100 lung cancer and 100 control subjects), yielding 91% sensitivity and 80% specificity in a 20-fold cross-validation. Our results clearly indicate that the proposed clinical model is suitable for non-invasive early lung cancer diagnosis, and is indifferent to lung cancer stage and histological type.




Similar content being viewed by others
Abbreviations
- AUC:
-
Area under the curve
- COPD:
-
Chronic obstructive pulmonary disease
- CV:
-
Cross-validation
- LDCT:
-
Low-dose computed tomography
- MAP:
-
Metabolic activity profile
- OXPHOS:
-
Oxidative phosphorylation
- ROC:
-
Receiver-operating characteristic
- SCLC:
-
Small cell lung cancer
- SVM:
-
Support vector machine
- USPSTF:
-
US Preventive Services Task Force
References
Dela Cruz CS, Tanoue LT, Matthay RA (2011) Lung cancer: epidemiology, etiology, and prevention. Clin Chest Med 32:605–644. https://doi.org/10.1016/j.ccm.2011.09.001
Howlader N, Noone A, Krapcho M et al (2016) SEER Cancer Statistics Review 1975–2013. National Cancer Institute; Bethesda, MD. https://seer.cancer.gov/csr/1975_2013/. Accessed 4 May 2017
Smith RA, Cokkinides V, Brooks D et al (2011) Cancer screening in the United States, 2011. CA Cancer J Clin 61:8–30. https://doi.org/10.3322/caac.20096
Moyer VA (2014) Screening for lung cancer: U.S. preventive services task force recommendation statement. Ann Intern Med 160:330–338. https://doi.org/10.7326/M13-2771
Rampinelli C, De Marco P, Origgi D et al (2017) Exposure to low dose computed tomography for lung cancer screening and risk of cancer: secondary analysis of trial data and risk-benefit analysis. BMJ 356:j347. https://doi.org/10.1136/bmj.j347
Marshall HM, Bowman RV, Yang IA et al (2013) Screening for lung cancer with low-dose computed tomography: a review of current status. J Thorac Dis 5:S524–S539. https://doi.org/10.3978/j.issn.2072-1439.2013.09.06
Pinsky PF, Kramer BS (2015) Lung cancer risk and demographic characteristics of current 20–29 pack-year smokers: implications for screening. J Natl Cancer Inst 107:djv226. https://doi.org/10.1093/jnci/djv226
Rodriguez-Roisin R, Soriano JB (2008) Chronic obstructive pulmonary disease with lung cancer and/or cardiovascular disease. Proc Am Thorac Soc 5:842–847. https://doi.org/10.1513/pats.200807-075TH
Siravegna G, Marsoni S, Siena S, Bardelli A (2017) Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol 14:531–548. https://doi.org/10.1038/nrclinonc.2017.14
Aravanis AM, Lee M, Klausner RD (2017) Next-generation sequencing of circulating tumor DNA for early cancer detection. Cell 168:571–574. https://doi.org/10.1016/j.cell.2017.01.030
Fujii T, Barzi A, Sartore-Bianchi A et al (2017) Mutation-enrichment next-generation sequencing for quantitative detection of KRAS mutations in urine cell-free DNA from patients with advanced cancers. Clin Cancer Res 23:3657–3666. https://doi.org/10.1158/1078-0432.CCR-16-2592
Li X, Hayward C, Fong P-Y et al (2013) A blood-based proteomic classifier for the molecular characterization of pulmonary nodules. Sci Transl Med 5:207ra142. https://doi.org/10.1126/scitranslmed.3007013
Lanman RB, Mortimer SA, Zill OA et al (2015) Analytical and clinical validation of a digital sequencing panel for quantitative, highly accurate evaluation of cell-free circulating tumor DNA. PLoS One 10:e0140712. https://doi.org/10.1371/journal.pone.0140712
Brock G, Castellanos-Rizaldos E, Hu L et al (2015) Liquid biopsy for cancer screening, patient stratification and monitoring. Transl Cancer Res 4:280–290. https://doi.org/10.3978/j.issn.2218-676X.2015.06.05
Vachani A, Hammoud Z, Springmeyer S et al (2015) Clinical utility of a plasma protein classifier for indeterminate lung nodules. Lung 193:1023–1027. https://doi.org/10.1007/s00408-015-9800-0
Whitney DH, Elashoff MR, Porta-Smith K et al (2015) Derivation of a bronchial genomic classifier for lung cancer in a prospective study of patients undergoing diagnostic bronchoscopy. BMC Med Genom 8:18. https://doi.org/10.1186/s12920-015-0091-3
Anderson D, Najafzadeh M, Gopalan R et al (2014) Sensitivity and specificity of the empirical lymphocyte genome sensitivity (LGS) assay: implications for improving cancer diagnostics. FASEB J 28:4563–4570. https://doi.org/10.1096/fj.14-254748
Pantel K, Alix-Panabières C (2013) Real-time liquid biopsy in cancer patients: Fact or fiction? Cancer Res 73:6384–6388. https://doi.org/10.1158/0008-5472.CAN-13-2030
Hiley CT, Le Quesne J, Santis G et al (2016) Challenges in molecular testing in non-small-cell lung cancer patients with advanced disease. Lancet 388:1002–1011. https://doi.org/10.1016/S0140-6736(16)31340-X
Bettegowda C, Sausen M, Leary R (2014) Detection of circulating tumor DNA in early-and late-stage human malignancies. Sci Transl 6:224ra24. https://doi.org/10.1126/scitranslmed.3007094
Vander Heiden MG, Cantley LC, Thompson CB (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324:1029–1033. https://doi.org/10.1126/science.1160809
Kaelin WG Jr, Thompson CB (2010) Q&A: cancer: clues from cell metabolism. Nature 465:562–564. https://doi.org/10.1038/465562a
DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB (2008) The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab 7:11–20. https://doi.org/10.1016/j.cmet.2007.10.002
MacIver NJ, Jacobs SR, Wieman HL et al (2008) Glucose metabolism in lymphocytes is a regulated process with significant effects on immune cell function and survival. J Leukoc Biol 84:949–957. https://doi.org/10.1189/jlb.0108024
Fox C, Hammerman P, Thompson C (2005) Fuel feeds function: energy metabolism and the T-cell response. Nat Rev Immunol 5:844–852. https://doi.org/10.1038/nri1710
Michalek RD, Rathmell JC (2010) The metabolic life and times of a T-cell. Immunol Rev 236:190–202. https://doi.org/10.1111/j.1600-065X.2010.00911.x
Pearce E (2010) Metabolism in T cell activation and differentiation. Curr Opin Immunol 22:314–320. https://doi.org/10.1016/j.coi.2010.01.018
Chang CH, Curtis JD, Maggi LB Jr et al (2013) Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell 153:1239–1251. https://doi.org/10.1016/j.cell.2013.05.016
Dietl K, Renner K, Dettmer K et al (2010) Lactic acid and acidification inhibit TNF secretion and glycolysis of human monocytes. J Immunol 184:1200–1209. https://doi.org/10.4049/jimmunol.0902584
Jellusova J, Cato MH, Apgar JR et al (2017) Gsk3 is a metabolic checkpoint regulator in B cells. Nat Immunol 18:303–312. https://doi.org/10.1038/ni.3664
Patsoukis N, Bardhan K, Chatterjee P et al (2015) PD-1 alters T-cell metabolic reprogramming by inhibiting glycolysis and promoting lipolysis and fatty acid oxidation. Nat Commun 6:6692. https://doi.org/10.1038/ncomms7692
Yang Z, Matteson EL, Goronzy JJ, Weyand CM (2015) T-cell metabolism in autoimmune disease. Arthritis Res Ther 17:29. https://doi.org/10.1186/s13075-015-0542-4
Beezhold K, Byersdorfer CA (2018) Targeting immuno-metabolism to improve anti-cancer therapies. Cancer Lett 414:127–135. https://doi.org/10.1016/J.CANLET.2017.11.005
Chimenti MS, Triggianese P, Conigliaro P et al (2015) The interplay between inflammation and metabolism in rheumatoid arthritis. Cell Death Dis 6:e1887. https://doi.org/10.1038/cddis.2015.246
Dunn GP, Bruce AT, Ikeda H et al (2002) Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol 3:991–998. https://doi.org/10.1038/ni1102-991
Swann JB, Smyth MJ (2007) Immune surveillance of tumors. J Clin Invest 117:1137–1146. https://doi.org/10.1172/JCI31405
Dunn GP, Old LJ, Schreiber RD (2004) The three Es of cancer immunoediting. Annu Rev Immunol 22:329–360. https://doi.org/10.1146/annurev.immunol.22.012703.104803
Michalek RD, Gerriets VA, Jacobs SR et al (2011) Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol 186:3299–3303. https://doi.org/10.4049/jimmunol.1003613
Pearce E, Poffenberger M, Chang C (2013) Fueling immunity: insights into metabolism and lymphocyte function. Science 342:1242454. https://doi.org/10.1126/science.1242454
Pedregosa F, Varoquaux G, Gramfort A et al (2011) Scikit-learn: machine learning in Python. J Mach Learn Res 12:2825–2830
Weissferdt A, Moran CA (2014) Reclassification of early stage pulmonary adenocarcinoma and its consequences. J Thorac Dis 6:S581–S588. https://doi.org/10.3978/j.issn.2072-1439.2014.07.41
Goldstraw P, Ball D, Jett JR et al (2011) Non-small-cell lung cancer. Lancet 378:1727–1740. https://doi.org/10.1016/S0140-6736(10)62101-0
Siegel RL, Miller KD, Jemal A (2015) Cancer statistics, 2015. CA Cancer J Clin 65:5–29. https://doi.org/10.3322/caac.21254
Sekine Y, Katsura H, Koh E et al (2012) Early detection of COPD is important for lung cancer surveillance. Eur Respir J 39:1230–1240. https://doi.org/10.1183/09031936.00126011
Acknowledgements
The authors wish to thank the principal investigators, Prof. Fernando Patolsky, Prof. Nadir Arber, Dr. Mirjana Wollner, and Dr. Ran Kremer; the coordinators of this study, including multi-site coordinator Janna Bronstein, for patient recruitment, collection of specimens, and clinical data; Dr. Sara Bar Yehuda for regulatory support; Dr. Uri Itay and Chaim Singal for their help in the statistical analysis of the data; and furthermore, all the donor patients for their contribution to this clinical study.
Funding
This work was supported by Savicell Diagnostics and by Grants from the Israel Science Foundation (ISF) through the Legacy Program.
Author information
Authors and Affiliations
Contributions
FP conceived and designed the experiments and assisted in manuscript preparation. HPS and RT designed and performed preliminary pre-clinical studies and helped analyzing preliminary data. ST, ED, GD, and IA synchronized the study, helped analyzing data, and assisted in manuscript preparation. SA, CB, TS, and AL performed clinical testing, assisted in data analysis, and assisted in manuscript preparation. ST and ED developed classification model and performed statistical analysis. YA, SS, and MS assisted in clinical sample collection, clinical study coordination, and manuscript preparation.
Corresponding author
Ethics declarations
Conflict of interest
Fernando Patolsky received consulting fees from Savicell Diagnostics. Shoval Tirman, Aviv Lutaty, Tali Scheinmann, Eyal Davidovits, Irit Arbel, and Giora Davidovits are employed by Savicell Diagnostics. Shirley Abramovitch and Cynthia Botbol were employed by Savicell Diagnostics. Eyal Davidovits, Irit Arbel, and Giora Davidovits are officers in Savicell Diagnostics. Eyal Davidovits and Giora Davidovits are on the board of directors of Savicell Diagnostics. Yochai Adir, Shoval Tirman, Shirley Abramovitch, Cynthia Botbol, Aviv Lutaty, Tali Scheinmann, Eyal Davidovits, Irit Arbel, Giora Davidovits, and Fernando Patolsky own stock and/or options in Savicell Diagnostics’ parent company. The other authors declare that they have no conflict of interest.
Ethical approval and ethical standards
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committees and with the 1964 Helsinki declaration and its later amendments. Institutional review board approval numbers are: 0105-13-CMC for Carmel Medical Center, Haifa; 0274-15-RMB for Rambam Medical Center, Haifa; and 0009-13-TLV for Sourasky Medical Center, Tel Aviv.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Adir, Y., Tirman, S., Abramovitch, S. et al. Novel non-invasive early detection of lung cancer using liquid immunobiopsy metabolic activity profiles. Cancer Immunol Immunother 67, 1135–1146 (2018). https://doi.org/10.1007/s00262-018-2173-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00262-018-2173-5