Abstract
Early, efficient Parkinson’s disease (PD) tests may facilitate pre-symptomatic diagnosis and disease-modifying therapies. Here we report elevated levels of PD-specific transfer RNA fragments carrying a conserved sequence motif (RGTTCRA-tRFs) in the substantia nigra, cerebrospinal fluid and blood of patients with PD. A whole blood qPCR test detecting elevated RGTTCRA-tRFs and reduced mitochondrial-originated tRFs (MT-tRFs) segregated pre-symptomatic patients with PD from controls (area under the receiver operating characteristic curve (ROC-AUC) of 0.75 versus 0.71 based on traditional clinical scoring). Strengthening PD relevance, patients carrying PD-related mutations presented higher blood RGTTCRA-tRFs/MT-tRFs ratios than mutation-carrying non-symptomatic controls, and RGTTCRA-tRF levels decreased in patients’ blood after deep brain stimulation. Furthermore, RGTTCRA-tRFs complementarity to ribosomal RNA and the translation-supporting LeuCAG3-tRF might aggravate PD via translational inhibition, as reflected by disrupted ribosomal association of RGTTCRA-tRFs in depolarized neuroblastoma cells. Our findings show tRF involvement in PD and suggest a potential simple and safe blood test that may aid clinicians in pre-symptomatic PD diagnosis after validation in larger independent cohorts.
Similar content being viewed by others
Main
Parkinson’s disease (PD) is the second most prevalent neurodegenerative disease1. PD poses major clinical, social and financial burdens to society2 and is characterized by progressive movement disabilities, tremors and cognitive impairments. These symptoms result from premature death of dopaminergic neurons, especially in the substantia nigra pars compacta (SN)3, that are caused by oxidative and nitrative stress3,4,5,6, enhanced mitophagy7, impaired autophagy8, neuroinflammation and dopaminergic/cholinergic imbalance9.
PD diagnosis often follows considerable neuronal damage manifested as severe motor impairments, such as bradykinesia, rigidity and tremors10. However, earlier symptoms, including smell loss and rapid eye movement sleep disorders, may appear years beforehand. Molecular changes characteristic of this early disease phase may constitute a basis for a pre-symptomatic diagnostic test. Such a test should be cost-effective, rapid and capable of detecting changes in biomarker levels at the earliest disease stages, confirming true-positive cases and minimizing false-positive ones. Recent PD diagnostic tests have focused on elevated cerebrospinal fluid (CSF) levels of the α-synuclein (α-Syn) protein or reduced blood mitochondrial DNA as biomarkers11,12,13,14. However, CSF sampling is invasive; purification and detection of α-Syn are cumbersome and insufficiently sensitive15; and measurements of specific proteins show high inter-individual variability. In contrast, quantification of multiple molecules, rather than one, can improve signal-to-noise ratios and overcome the variability between patients16,17,18,19,20,21. Attempting to prevent invasive tests, certain blood molecules were proven to reflect cerebral pathology22. Ideally, an easy, safe and affordable diagnosis should be based on multiple highly sensitive and specific blood biomarkers23.
Transfer RNA fragments (tRFs) are 16–50 nucleotide (nt)-long, non-coding RNAs originating from multiple nuclear or mitochondrial transfer RNA (tRNA) genes. Several enzymes cleave tRNAs to yield tRFs: angiogenin (Ang) generates 5′-half and 3′-half tRFs, whereas Ang, Dicer24,25 and other nucleases yield 5′-tRFs, i-tRFs and 3′-tRFs26. Because tRFs harbor repetitive sequence motifs inherited from their parental tRNAs, changes can be detected in an entire family of closely related transcripts, albeit of different origins and types, produced by different endonucleases20,21,27, and originating from either nuclear or mitochondrial genomes. Correspondingly, tRFs may reflect transcriptional changes or malfunctions in both the nucleus and the mitochondria27, whose DNA is known to be damaged in PD14,28,29. Intriguingly, altered Ang levels, Ang mutations and Ang-produced tRFs may exert both PD-protective and cytotoxic roles30,31,32,33. Furthermore, blood cell tRF levels present diagnostic value in various diseases, including ischemic stroke34, amyotrophic lateral sclerosis35, epilepsy36 and cellular or organismal stress26,34. Accordingly, tRFs emerge as the perfect candidates for a blood-based PD biomarker.
To challenge the hypothesis that multiple tRF-derived sequences could yield informative PD biomarkers, we searched for tRF families that share common sequence motifs whose levels are changed in the brain, blood and/or CSF of patients with PD—all biofluids known to reflect cerebral pathologies, including PD22,37. Using different datasets that account for distinct biofluids and stages of the disease allowed us to study the PD links of those tRFs whose levels had been altered in unrelated cohorts of different population origins.
Results
tRF levels in CSF show PD-specific changes
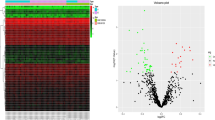
To seek PD-characteristic tRF profiles, we analyzed a short RNA sequencing (RNA-seq) dataset (phs.000727)38 consisting of postmortem (PM) CSF samples from 46 patients with PD, from 53 patients with Alzheimer’s disease (AD) and from 53 healthy controls (15, 27 and 24 females, respectively; Fig. 1a, Extended Data Fig. 1a,b and Supplementary Table 1a,b). Accounting for patients’ age, sex and PM interval (Extended Data Fig. 2a; edgeR; Methods), we identified elevated levels of nuclear-originated tRFs in patients with PD compared to controls (N-tRFs; average levels in control (Ctrl) and PD: 980,165 and 990,387 counts per million (CPM), respectively). All but one of these were 3′-tRFs or i-tRFs that were over 35 nt long, excluding them from functioning as microRNAs39 (Fig. 1b and Extended Data Fig. 2b,c). We also detected reduced levels of mitochondrial-originated tRFs (MT-tRFs; average levels in Ctrl and PD: 19,834 and 9,612 CPM, respectively; Fig. 1b)14,28, most of which are long 3′-tRFs (Extended Data Fig. 2d,e). The PD-elevated N-tRFs and reduced MT-tRFs phenomena were consistent in both sexes (Extended Data Fig. 2b,c) but were absent in AD (Fig. 1c), reflecting non-random PD-altered CSF tRF profiles37.
a, Schematic representation of N-tRFs and MT-tRFs in PM CSF and SN samples. b, Levels of MT-tRFs (left; green background) and N-tRFs containing and lacking the RGTTCRA motif (right and middle; blue background) in PD CSF. Each dot represents a tRF. x: log2(fold change (FC)) of PD versus Ctrl levels. y: −log10(FDR-adjusted P value). Horizontal blue line: FDR = 0.05. Vertical dashed lines: log2(FC) > 1 or log2(FC) < −1. Dot colors: tRF lengths. c, As described in b for AD versus Ctrl. d, MT-tRFs decrease and elevation of RGTTCRA-tRFs associates with Lewy body scores in the SN. x axis: log2(FC) for high versus low Lewy body score (that is, prevalence and localization of brain Lewy bodies). y axis: −log10(FDR-adjusted P values). Colors are as in b.
N-tRFs elevated in PD CSF share a common RGTTCRA motif
Pursuing PD RNA biomarkers, we sought common motifs shared by the elevated N-tRFs using the Multiply Elicited Motif Entities (MEME) tool40. This tool detected multiple tRFs sharing the sequence [A/G]GTTC[A/G]A (RGTTCRA-tRFs; Fig. 1b and Extended Data Fig. 1c), most of which were over 30 bases long (average levels: Ctrl = 79,901, PD = 92,836, AD = 83,068, CPM for RGTTCRA-tRFs >30 bases). Binomial tests confirmed that RGTTCRA-tRFs are mostly elevated and MT-tRFs are mostly reduced also when analyzing each sex separately (Extended Data Fig. 2 and supplementary information for Extended Data Fig. 2). Notably, RGTTCRA-tRFs were derived from numerous nuclear-originated tRNA genes, none of which was downregulated in the CSF of patients with PD, and all of the CSF-elevated tRFs lacking this motif were shorter than 30 bases (Fig. 1b). Together, this indicates that the shared motif embedded in RGTTCRA-tRFs might reflect disease-related features (Supplementary Table 2 lists all of the tRNA genes carrying the PD motif).
Parallel analysis comparing patients with AD to controls in the same dataset revealed unchanged profiles of MT-tRFs and RGTTCRA-tRFs (Fig. 1c). Moreover, the median levels of RGTTCRA-tRFs longer than 30 bases were higher in patients with PD than in either healthy controls or patients with AD (P < 0.038 and P < 0.062, respectively, Dunnett test, false discovery rate (FDR)). The PD-specific accumulation of RGTTCRA-tRFs and decrease of MT-tRFs in CSF identified both of these subfamilies as promising PD biomarker candidates.
RGTTCRA-tRFs co-accumulate with Lewy bodies in the SN of patients with PD
To seek pathological implications of RGTTCRA-tRF accumulation, we conducted small RNA-seq of SN from eight PM patients with PD sourced from the Netherlands Brain Bank (NBB) (Supplementary Figs. 1a,b and 2h and Supplementary Table 3; data deposited in GSE256334). Supporting our CSF findings, elevation of RGTTCRA-tRFs (average levels = 91,856 CPM) and reduction of MT-tRFs (average levels = 54,123 CPM) accompanied higher Braak Lewy body scores in patients with PD (Fig. 1d).
Ang is sufficient but not essential for production of RGTTCRA-tRFs
Seeking the cleavage enzymes responsible for the altered RGTTCRA-tRFs and MT-tRFs (Fig. 2a), we analyzed a dataset of Ang overexpression in HEK293T and knockout in U2OS cells (GSE130764; Supplementary Table 4)41. Ang overexpression elevated the percentage of RGTTCRA-tRFs, but not of MT-tRFs, in HEK293 cells (P < 0.0093 and P < 0.375, respectively, t-test, FDR; Fig. 2b–d). However, Ang knockout had no overt effects on these tRF families (Extended Data Fig. 3a,b and supplementary information for Extended Data Fig. 3). Therefore, Ang emerged as sufficient but not essential for producing RGTTCRA-tRFs or MT-tRFs.
a, Scheme of tRNA cleavage by Ang and Dicer. b, tRFs were quantified in Ctrl and Ang-overexpressing (OE) HEK293T cells. c,d, RGTTCRA-tRF (c, purple) and MT-tRF (d, green) percentages in Ang-OE versus Ctrl. Each dot represents a biological replicate (n = 3 in each condition). **P < 0.0093 and P < 0.375 for c and d, respectively, two-sided t-test, FDR correction. e, Numbers of blood samples from live controls and early PD patients with and without PD-related mutations (PPMI) and PM advanced PD patients (NBB). f,g, Percentage of RGTTCRA-tRFs (f, purple) and MT-tRFs (g, green) in Ctrl and PD samples (PPMI, ‘Early’; Idiopathic, carrying no PD-related mutations; Genetic, carrying LRRK2, GBA or SNCA mutations; NBB, ‘Advanced’). Each dot represents a patient (n = 133, 252, 16, 55, 16 and 21; from leftmost box plot to rightmost one). For RGTTCRA-tRFs and MT-tRFs in advanced patients: ***P < 0.00015 and *****P < 8 × 10−11, respectively, two-way ANOVA. For MT-tRFs in early Genetic patients: #P < 0.057, two-sided Mann–Whitney test, FDR correction. h, RGTTCRA-tRFs/MT-tRFs ratio in the above six groups. *P < 0.02 and ****P < 0.00044 for early Idiopathic and Genetic patients, respectively, and P < 0.0013 for PD versus Ctrl in all groups. Two-way ANOVA. All box plots in this figure are defined so that the central line of the box plot represents the median; the lower and upper box bounds represent 0.25 and 0.75 quantiles; and lower and upper whisker bounds represent 0 and 1 quantiles.
Blood RGTTCRA-tRFs and MT-tRFs reflect disease features in early and advanced PD patients
To test the utility of RGTTCRA-tRF and MT-tRF levels as diagnostic biomarkers, we performed small RNA-seq of NBB PM blood samples from patients with PD and controls (Ctrl, n = 16 and PD, n = 21; Fig. 2e and Supplementary Table 5; data deposited in GSE256334) and used Parkinson’s Progression Markers Initiative (PPMI) data (Supplementary Table 6) from idiopathic non-deep brain stimulation (DBS)-treated patients from timepoint V08 (2 years after first evaluation; Idiopathic; PD, n = 252 and Ctrl, n = 133) and the latest timepoint available for PD-related mutation carriers with or without symptoms (Genetic; PD, n = 55 and Ctrl, n = 16).
Mutation-carrying living PPMI PD patients showed a trend of reduction in blood MT-tRF levels compared to unaffected mutation-carrying controls (P < 0.057, Mann–Whitney, FDR; Fig. 2g). In comparison, PM blood samples from advanced NBB PD patients presented elevated RGTTCRA-tRFs and MT-tRFs (P < 0.00015 and P < 8 × 10−11, respectively, ANOVA; ‘Advanced’; Fig. 2f,g).
The observed PM elevation in blood MT-tRF levels contrasts with the MT-tRF reduction seen in the PD CSF and SN (Fig. 1b,d) and may reflect mitochondrial degradation in PM blood, which yields higher levels of mitochondrial RNA fragments42. Indeed, although levels of RGTTCRA-tRFs were not affected by sample RNA integrity number (RIN), MT-tRF levels were negatively correlated with the sample’s RIN (MT-tRFs: r = −0.4, P < 0.025; RGTTCRA-tRFs: r = −0.14, P < 0.4; FDR, Spearman correlation), indicating that the elevated levels stem from the PM status rather than the PD status. Supporting this notion, SN tissues revealed reduced mitochondrial tRNA levels, which continued declining with disease duration (r = 0.4, P < 0.05, Spearman correlation; PD versus Ctrl P < 0.039, Mann–Whitney test; Extended Data Fig. 3c).
To develop a biomarker assay that can easily compare among patients regardless of the need to account for RNA concentrations and library depth, we calculated the ratio between RGTTCRA-tRFs and MT-tRFs. This ratio was elevated in PD versus Ctrl (Fig. 2h; P < 0.0013, ANOVA), especially in early mutation-carrying patients (Early Idiopathic patients P < 0.02, Early Genetic patients P < 0.00044, ANOVA). Strengthening the notion that blood tRFs have the capability of capturing inter-individual variations, we found that blood tRF levels create a patient-specific fingerprint that maintains stability over time, positioning them as promising candidates for biomarkers (Supplementary Information: ‘Blood tRF levels create a patient-specific fingerprint’).
Feature selection-based in silico tRF probes outperform clinical measurements in separating prodromal PD from control patients
We next challenged the value of the RGTTCRA-tRFs/MT-tRFs ratio for pre-symptomatic PD detection. To do so, we compared prodromal PPMI patients (showing pre-symptomatic PD signs, albeit without a clinical PD diagnosis) at the earliest tested timepoint (n = 60) to matched healthy controls (n = 128) (Fig. 3a and Supplementary Table 7). A subset of 60 controls optimally matched to the prodromal patients was formed based on sex, age and collection batch of patients.
a, GBM machine learning (ML) algorithm segregated PPMI prodromal patients from controls using the ratio between RGTTCRA-tRFs (GGTCCCTGGTTCAA sequence) and MT-tRFs (TAACTTAGCATTAACCTTTTAA sequence), compared to clinical measurements. b, ROC of optimally matched prodromal patients and controls (n = 60 of each), using tRF-score (orange) or clinical UPDRS and H&Y measurements (turquoise) and mixed labels combining tRFs and clinical measurements (gray). c, Density plot of AUCs from 10,000 training events, each using a different pair of random 14-nt and 22-nt motifs (without RGTTCRA motif) to calculate ‘tRF-score’ and GBM-based AUCs. Red area: s.d. d, tRF-score in control (gray) and prodromal patients (orange) of different ethnic and genetic backgrounds. Dots represent patients. Prodromal versus Ctrl P < 2 × 10−10, two-way ANOVA. e, As in b for a holdout validation sample (33 prodromal and 33 Ctrl). f, GBM algorithm trained in b applied to holdout validation sample (n = 33 in each group). x: true classification. y: GBM-based prediction of prodromal patient diagnosis. ***P < 0.0004, two-sided t-test. g, tRF-score in control (gray) and 21 prodromal patients, later diagnosed as PD (orange). *****P < 1.1 × 10−5, two-sided t-test, FDR. h, As in f for UPDRS scores. ***P < 7 × 10−3, two-sided t-test, FDR. i, Cohorts used for the qPCR test. j, RGTTCRA/MT-tRF qPCR-based separation in fresh blood samples of PD or trauma patients and controls (10 of each). PD versus Ctrl: *P < 0.05, PD versus trauma: *P < 0.013, Dunnett test. k, As in i using 23 SN samples from the NIH NeuroBioBank. *P < 0.0452, two-sided t-test. All box plots in this figure are defined so that the central line of the box plot represents the median; the lower and upper box bounds represent 0.25 and 0.75 quantiles; and lower and upper whisker bounds represent 0 and 1 quantiles.
To create a primer-based separation reflecting the variability between controls and prodromal PD patients, we sought two single in silico polymerase chain reaction (PCR) primer pairs that can recognize numerous RGTTCRA-tRFs or MT-tRFs. We designed quantitative PCR (qPCR)-suitable 14-nt-long sequences that include the PD motif and are shared among at least 100 different RGTTCRA-tRFs (Methods). For every patient, we summed the counts of all RGTTCRA-tRFs, including each 14-nt sequence, and selected the one that led to the largest median difference between PPMI prodromal patients and healthy controls (GGTCCCTGGTTCAA, shared among 285 RGTTCRA-tRFs from different tRNAs of distinct chromosomal origins, average levels of 550 CPM; P < 0.013, Mann–Whitney). Next, we similarly selected a sequence shared between the maximum number of MT-tRFs that maximizes the median difference between prodromal and controls and minimized the standard deviation within each group (TAACTTAGCATTAACCTTTTAA, shared among 106 Lys tRNA-originated MT-tRFs, average levels of 90 CPM).
For each patient, we divided the sum of RGTTCRA-tRF counts (all carrying the GGTCCCTGGTTCAA sequence) by the sum of MT-tRF counts (all carrying the TAACTTAGCATTAACCTTTTAA sequence) to yield an RGTTCRA/MT score. The RGTTCRA/MT score of each control or prodromal patient from a matched pair was then divided by the pair’s mean to yield a normalized score (hereafter ‘tRF-score’). Next, we employed a gradient boosted machine learning (GBM) classification algorithm (five cross-validations; Methods) to subgroup the samples into prodromal PD patients and controls, considering their ethnicity, by clinical measurements of Unified PD Rating Scale (UPDRS) motor scores and Hoehn and Yahr (H&Y) scores or by the tRF-score described above. This resulted in areas under the curve (AUCs; representing the ability to segregate the patients into control and prodromal groups) of 0.73 for the clinical measurement (specificity = 0.78, sensitivity = 0.583, positive predictive value (PPV) = 0.761, negative predictive value (NPV) = 0.61) compared to an AUC of 0.86 (specificity = 0.867, sensitivity = 0.7, PPV = 0.84, NPV = 0.743) for the tRF-score (Fig. 3a,b). In comparison, classification using mixed labels (null) in both clinical and tRF measurements yielded a non-significant AUC value of 0.43. Further validation using 10,000 permutations, each time sampling 60 different controls, confirmed these results, as did a logistic regression algorithm replacing GBM (receiver operating characteristic (ROC)-AUC of 0.83 compared to 0.70 using clinical measurements; Extended Data Fig. 4a).
To test if our findings reflect the biological role of these motifs rather than training of the algorithm, we ran the exact same GBM-based process 10,000 times on ratios between random motifs (lacking the RGTTCRA sequence and appearing in at least the same number of tRFs as the RGTTCRA motifs and the MT motifs; Methods). The mean AUC received for the ratio between random motifs was 0.768 (s.d. = 0.0537), and 99% of the AUCs were lower than 0.81 (Fig. 3c). These findings support the notion that the AUC resulting from the tRF-score reflects a biological rather than a mere training phenomenon.
PD symptoms often reflect patients’ genetics and ethnic origins. Therefore, we tested the ability to segregate patients to prodromal and control groups based on their genetic and ethnic backgrounds. The tRF-score enabled segregating all subpopulations (Fig. 3d; P < 2 × 10−10, ANOVA), even when examining the non-normalized ratio (Extended Data Fig. 4b; P < 5 × 10−6, ANOVA). Thus, two-thirds of prodromal patients scored as 6 or higher, whereas two-thirds of controls scored lower than 6 (P < 4 × 10−5, chi-square test). An exception was Black control and prodromal patients who had similar tRF-scores. However, their UPDRS scores revealed the same trend (Extended Data Fig. 4c), potentially suggesting that our test reflected real-life clinical measurements in these patients as well. Nonetheless, although the tRF-score measurement was similar between prodromal and diagnosed PD patients, the UPDRS score of prodromal patients was akin to that of controls and differed dramatically from that of patients with PD (Extended Data Fig. 4d). Further indicating a link between our two motifs and the clinical measurements, the tRF-scores of the optimally matched prodromal and control patients presented positive correlation with the total UPDRS score (r = 0.27, P < 0.01; FDR, Spearman correlation; Extended Data Fig. 4e) and with the scores of subsections II and III of the UPDRS test (I: r = 0.19, P < 0.06; II: r = 0.245, P < 0.0163; III: r = 0.318, P < 0.004; FDR, Spearman correlation; Extended Data Fig. 4f).
The tRF-score strengthens clinical measurements in predicting PD pre-symptomatically
Next, we tested the GBM algorithms trained on the optimally matched data (60 prodromal, 60 controls) on the fully matched data (60 prodromal, 128 controls) (Methods). Both algorithms, whether trained on the tRF-score or on clinical parameters, presented far better classification abilities than random classification (Extended Data Fig. 4g; P < 5 × 10−14, P < 3 × 10−7, chi-square test, FDR). However, the tRF-based measurements performed better than the clinical ones (P < 0.0081, chi-square test). To re-challenge these observations, we tested the tRF-score-based algorithm on a holdout validation sample, including 33 prodromal samples and 33 matched control samples consisting of the second-earliest timepoint of the patients used for cross-validation (discovery sample) (Methods and Supplementary Table 7). The algorithm successfully distinguished prodromal patients from controls (P < 0.0004, t-test; AUC of 0.75, confidence interval (CI) = 0.63–0.86, specificity = 0.727, sensitivity = 0.606, PPV = 0.69, NPV = 0.649; Fig. 3e,f), re-demonstrating the algorithm’s efficacy and the ability of the tRF-score to identify prodromal patients. In comparison, testing the algorithm trained on the clinical measurement on the holdout validation sample yielded a weaker separation between control and prodromal patients (AUC of 0.71, CI = 0.65–0.78, specificity = 0.523, sensitivity = 0.777, PPV = 0.58, NPV = 0.734; Methods). Together, the tRF-score performed at least as well as the clinical measurement and, in certain cases, better than the clinical tests, both on the cross-validated discovery sample and on the holdout validation sample.
In subsequent tests, PD diagnosis was established for 21 of those 60 studied prodromal PPMI patients (with the rest yet undiagnosed). Comparing the tRF and UPDRS scores of these 21 patients to those of controls yielded a more significant separation by the tRF-score (P < 1.1 × 10−5 versus P < 7 × 10−3, t-test, FDR; Fig. 3g,h). Furthermore, 18 of the 21 patients whose PD diagnosis had been confirmed were ranked higher by their tRF-score than by their UPDRS score (Extended Data Fig. 4h), again emphasizing the advantage of using the dual tRFs measurement. In later stages of PD with overt motor symptoms (Supplementary Table 8), clinical measurements perfectly separated patients from controls (AUC of 1), which left no room for added value of other biomarkers. We conclude that the two-primer-based tRF-score emerges as a reliable PD biomarker already in the earliest disease stages and especially in combination with clinical or other available measurements.
A dual qPCR of RGTTCRA-tRFs and MT-tRFs segregated PD from controls in fresh blood and PM SN samples
To segregate patients with PD from controls based on a manual qPCR-based tRF test, we drew blood from 10 patients with PD, from 10 patients with head trauma and from 10 controls from the Jerusalem Shaare Zedek Medical Center (JLM cohort; Supplementary Table 9). We used the two probes described above (ΔCq = RGTTCRA-tRF − MT-tRF; Methods) and normalized the ΔΔCq separately for each sex (Methods and Extended Data Fig. 1j). Notably, patients with PD presented significantly higher ratios of RGTTCRA-tRFs/MT-tRFs than either controls or patients with trauma, which showed similar values (Fig. 3i,j; P < 0.05, P < 0.013, Dunnett test).
To further validate the capacity of our qPCR test to selectively amplify the RGTTCRA motif, we subjected the amplicons of two qPCR reactions (JLM cohort, one patient with PD and one patient with trauma) to small RNA-seq. In both samples, over 50% of the reads (compared to 0.3–10% in biological samples) were mapped to tRFs, all carrying the RGTTCRA motif, with most derived from the same tRNA region but from different tRFs (Extended Data Fig. 4i and Supplementary Table 10). These findings provide experimental evidence supporting the notion that our qPCR test primarily detects tRFs that include the RGTTCRA motif and can quantify their levels in patients’ blood.
To address the consistency of our findings in other tissues, we tested 17 PD SN samples (six females) and six matched control SN samples (three females) from the National Institutes of Health (NIH) NeuroBioBank collection (Supplementary Table 11). Because tRFs expressed in the brain are slightly different than those in the blood, we designed a brain-adapted RGTTCRA-tRF primer (CGGGTTCGATTCCC; Methods) to maximize the number of detected RGTTCRA-tRFs. Our qPCR assay identified higher RGTTCRA-tRFs/MT-tRFs ratios in PD than in Ctrl (Fig. 3i,k; P < 0.0452, t-test). Together, our findings in blood and brain demonstrated that the signal of motif-carrying tRFs identified in sequenced blood samples from prodromal and PD patients can be translated into simple dual qPCR blood tests.
RGTTCRA-tRFs show ‘dual-lock’ sequence complementarity to rRNAs and to the translation-essential LeuCAG3′ tRF
Although MT-tRF reduction reflects well-studied PD-related mitochondrial dysfunction28,29 that potentially leads to reduction in MT-tRFs, our finding of RGTTCRA-tRF accumulation might reflect biological activities accelerating PD progression. In this context, four different tRFs in Saccharomyces cerevisiae were shown to interrupt translation by interacting with ribosomal RNA (rRNA), in regions other than tRNA binding sites43. Interestingly, we found all four to include the PD motif. Furthermore, we identified a region complementary to the RGTTCRA-tRF sequence in the leucine tRNA-derived tRF (LeuCAG3′) that was reported to be essential for assembly of the translation complex, protein translation and cellular viability44 (Fig. 4a,b). Correspondingly, minimal energy assessment (RNAup)45 predicted a thermodynamically preferred interaction (total energy of −10.46 kcal mol−1) of RGTTCRA-tRFs with the ribosome-essential LeuCAG3′ tRF (Fig. 4b), far lower binding energy than all other 1,017 CSF-identified tRFs (P < 1 × 10−30, t-test; Fig. 4b).
a, RGTTCRA-tRFs (purple) can hybridize both with a ribosome-essential tRF (Ribo-essential, cyan) and with 18S and 28S rRNAs, producing a ‘dual-lock’ translational arrest. b, Left, total binding energy of the Ribo-essential tRF to all CSF tRFs lacking (gray) or carrying (purple) the RGTTCRA motif (n = 1,017 tRFs). *****P < 1 × 10−30, two-sided t-test. Right, predicted interaction between a RGTTCRA-tRF (purple) and the Ribo-essential tRF (cyan) with PD motif nucleotides marked (orange asterisks). c, Left, total binding energy of 18S and 28S regions carrying RGTTCRA motif-complementary sequences to all CSF tRFs lacking (gray) or carrying (purple) the RGTTCRA PD motif. *****P < 2 × 10−25, *****P < 3 × 10−67, two-sided t-test, FDR. Right, secondary structures of 18S and 28S rRNA showing RGTTCRA motif-complementary sequences (red). All box plots in this figure are defined so that the central line of the box plot represents the median; the lower and upper box bounds represent 0.25 and 0.75 quantiles; and lower and upper whisker bounds represent 0 and 1 quantiles.
Supporting the ribosome-binding prediction, we found sequences complementary to the PD motif that are not involved in a secondary structure in both the 18S and 28S rRNAs and are, hence, amenable to hybridization (Fig. 4a,c). Correspondingly, RGTTCRA-tRFs were predicted to bind more strongly to 18S and 28S rRNAs than all other CSF tRFs (P < 2 × 10−25, P < 3 × 10−67, t-test, FDR; Fig. 4c). The predicted binding of RGTTCRA-tRFs to both the LeuCAG3′ tRF and the rRNA may lead to a ‘dual-lock’ mechanism interrupting both initiation and execution of translational processes (Fig. 4a). Indeed, 3 h of translation-interfering arginine or leucine starvation in three human-originated cell lines46 led to enrichment of ribosome-bound RGTTCRA-tRFs (P < 0.023, P < 0.027, Dunnett test; Extended Data Fig. 5a and supplementary text).
We next employed Förster resonance energy transfer (FRET) fluorescence lifetime imaging (FLIM)47 with pulsed interleaved excitation (PIE) (Extended Data Fig. 5b,c) to assess the proximities between RGTTCRA-tRF (that is, donor fluorophore-labeled RGTTCRA-tRF probe) and ribosomes (that is, acceptor fluorophore tagging an antibody to the ribosomal protein RPL24) (Extended Data Fig. 5c). The fluorescence lifetime (τ) of the acceptor fluorophore was longer after donor excitation (FRET-sensitized acceptor fluorescence) than after direct acceptor excitation (directly excited acceptor fluorescence; 1.36 ns versus 1.00 ns; Extended Data Fig. 5d). The delayed acceptor fluorescence after donor excitation may reflect a signature of FRET between the donor–acceptor pairs48 and, hence, close proximity (<10 nm) of RGTTCRA-tRFs with ribosomes (Extended Data Fig. 5d,f), supporting direct interaction between RGTTCRA-tRFs and the ribosome.
DBS is accompanied by reduced blood RGTTCRA-tRF levels
DBS of the subthalamic nucleus suppresses PD tremor symptoms through an incompletely understood mechanism of action49,50. To search for RGTTCRA-tRF links to the disease symptoms, we studied the effect of DBS on blood RGTTCRA-tRF levels in 19 DBS-treated patients with PD of the PPMI cohort carrying LRRK2 (ref. 51) or GBA mutations (Fig. 5a and Supplementary Table 12). Non-DBS-treated PD patient carriers of the same PD-causing mutations (n = 11) showed higher RGTTCRA-tRF levels than apparently healthy carriers of these mutations (n = 9; P < 0.027, ANOVA). In comparison, the DBS-treated mutation-carrying patients presented lower RGTTCRA-tRF levels than non-DBS-treated patients (P < 0.095, ANOVA; Fig. 5b), equivalent to the levels of the apparently healthy mutation-carrying controls (P < 0.6, ANOVA; Fig. 5b). MT-tRFs showed no significant difference between DBS-treated and the other groups (Extended Data Fig. 6a). The clinical capacity of DBS to effectively co-suppress blood RGTTCRA-tRF levels and tremor symptoms suggests that RGTTCRA-tRF blood levels may be related to the tremor profiles of PD patients.
a, Blood was drawn from controls and patients with PD with and without DBS from the PPMI and Soreq datasets. b, RGTTCRA-tRF levels in control carriers of PD-related mutations (gray) and PD carriers of mutated LRRK2 or GBA genes (blue) with and without DBS (red and black outlines), n = 9, 11, 19. y: percentage of RGTTCRA-tRFs from total tRF counts (rhombuses indicate mean). PD versus Ctrl *P < 0.027, PD versus DBS #P < 0.095, one-way ANOVA. c, Ang levels in blood samples of controls and patients with PD before and after DBS treatment, from the Soreq dataset (GSE23676), colors as in b. Dashed lines connect pre-DBS and post-DBS samples of each patient (six Ctrl, seven PD). PD pre-DBS versus DBS *P < 0.019, one-way ANOVA. d, Small RNA-seq and ribosome-bound RNA-seq fractions of depolarized SHSY-5Y cells (GSE155727) conducted at resting, immediately after depolarization (Dep) and 2 h pDP. e, RGTTCRA-tRF (y axis) fractions in each of the cases described in d (x axis). Four biological replicates in each condition. Cytosolic small RNA: resting versus Dep *P < 0.01, resting versus 2 h pDP *P < 0.015, Dep versus 2 h pDP ***P < 0.0001; ribosome fraction: 2 h pDP versus resting *P < 0.015, 2 h pDP versus Dep *P < 0.0065; two-way ANOVA. f, Total and ribosome-bound fractions of RGTTCRA-tRFs (purple), MT-tRFs (green) and all other tRFs (gray) in whole cells (left) and ribosome-bound fractions (right). Columns: samples and black lines represent mean RGTTCRA-tRF or MT-tRF percentages in samples. Reduction in MT-tRF in cytosolic small RNA 2 h pDP, P < 0.0033, two-way ANOVA. Three samples with altered ratios of RGTTCRA-tRFs and MT-tRFs 2 h pDP in ribosomal fractions (compared to mean distribution in resting cells), *P < 0.022, **P < 0.006, *P < 0.024, chi-square, FDR. All box plots in this figure are defined so that the central line of the box plot represents the median; the lower and upper box bounds represent 0.25 and 0.75 quantiles; and lower and upper whisker bounds represent 0 and 1 quantiles.
Seeking potential causes for the reduced percentage of RGTTCRA-tRFs out of total tRFs, and considering that DBS alters blood transcript profiles52 as well as the effect of Ang on RGTTCRA-tRF levels, we measured Ang levels in DBS-treated patients and controls. For this purpose, we used our microarray dataset, GSE23676 (ref. 52), consisting of blood taken from six controls and seven patients with PD (all males), where PD blood was drawn before DBS surgery or several months after DBS treatment (Fig. 5a, Soreq dataset; Methods, Extended Data Fig. 1k and Supplementary Table 13). Interestingly, Ang mRNA levels were similar in controls and patients with PD (before DBS) but decreased in six of the seven patients with PD after DBS (P < 0.019, ANOVA; Fig. 5c), possibly reflecting a limited post-DBS capacity to produce tRFs.
Depolarization of cultured cells reduces RGTTCRA-tRFs and impairs their ribosomal association
To examine the effects of electrical stimulation (such as DBS) on RGTTCRA-tRFs, we mined a short RNA-seq and ribosomal profiling dataset (GSE155727)53 from SHSY-5Y neuroblastoma cells harvested before, immediately after or 2 h after electrophysiological depolarization (2 h post depolarization (pDP); Fig. 5d and Supplementary Table 14). Immediately after depolarization, the cytosolic fraction of these cells showed reduced RGTTCRA-tRF shares, followed by re-elevation 2 h later (reminiscent of our findings in DBS-treated patients; Fig. 5e; resting versus depolarization, P < 0.01; resting versus 2 h pDP, P < 0.015; depolarization versus 2 h pDP, P < 0.0001, ANOVA). The re-elevation of RGTTCRA-tRFs share was accompanied by reduced cytosolic MT-tRFs share (Fig. 5f; P < 0.0033, ANOVA). Although depolarization did not immediately change the ribosomal-bound fraction of RGTTCRA-tRFs, this fraction was reduced 2 h later (Fig. 5e; 2 h pDP versus resting, P < 0.015; 2 h pDP versus depolarization, P < 0.0065, ANOVA). The distribution of tRFs to MT-tRFs, RGTTCRA-tRFs and all other tRFs was further altered 2 h pDP in the ribosome-bound fraction of three samples compared to the mean distributions of this fraction in resting cells (P < 0.006, P < 0.022, P < 0.024, chi-square, FDR; Fig. 5f).
Because RGTTCRA-tRFs mainly belong to the 3′ and i-tRF subtypes, we compared the fraction of RGTTCRA-tRFs in these specific subtypes and found the same association patterns as described above (Extended Data Fig. 6b and supplementary information for Extended Data Fig. 6). Although RGTTCRA-tRFs constituted a small portion of the total cytoplasmic short RNAs, their share of the ribosomal-associated RNAs was roughly 10-fold higher (P < 8 × 10−11, ANOVA; Fig. 5f). Our FRET findings, together with the selectively weakened ribosomal association of RGTTCRA-tRFs within 2 h after electrophysiological depolarization, suggest that the depolarizing qualities of DBS may impact the ribosomal association of RGTTCRA-tRFs. Altogether, RGTTCRA-tRFs and MT-tRFs showed selective and distinct co-regulation under electrophysiological stimuli at both the cellular and organismal levels.
Discussion
From our study, tRFs carrying a repetitive motif emerged as potentially suitable biomarkers that may constitute patient-specific ‘fingerprints’ and carry short conserved sequence motifs that enable single measurement of multiple tRFs. Intriguingly, we found that RGTTCRA-tRFs accumulate in the brain, CSF and blood of male and female patients with PD at diverse disease stages but not in matched controls or in patients with AD. Moreover, motif-carrying RGTTCRA-tRFs consistently showed linkage to PD symptoms and disease stages, and their levels were elevated in correlation with Lewy body scores in patients’ SN. Additionally, part of the identified RGTTCRA-tRFs stem from tRNAs that carry phenylalanine or cysteine amino acids, known to be the rate-limiting factors in the dopamine synthesis and in glutathione reductase antioxidant mechanism, respectively. Thus, shortage of these intact tRNAs (as they are enzymatically cut into the observed tRFs) may correspond to impaired dopamine synthesis54 or to processes that limit cellular antioxidation55. Compatible with the known mitochondrial damage in PD28 (which leads to general reduction in mitochondrial transcript levels), we further found reduced levels of MT-tRFs in the CSF and SN of idiopathic PD patients and in the blood of early PD patients carrying disease-related mutations.
Our segregation algorithm focused on prodromal patients rather than patients at later stages of PD because it is these early disease stages that call for biomarker development, especially as, in later stages, clinical measurements suffice to determine a clear diagnosis of PD. Correspondingly, the ratio of RGTTCRA-tRFs to MT-tRFs segregated early prodromal patients from healthy controls as well as or even better than those of the gold standard clinical UPDRS and H&Y measurements. This could indicate that the ratio of RGTTCRA-tRFs to MT-tRFs may serve as a potential diagnostic tool for the earliest stages of PD, especially in combination with other available pre-symptomatic detectors. To our knowledge, this evaluation, which is based on publicly available data sources and manual tests, is the first to identify the power of blood tRFs as PD biomarkers. Nonetheless, this cross-sectional study calls for longitudinal studies following patients throughout disease progression to offer insights into biomarker changes and treatment outcome.
As modern medicine aims to achieve patient-specific therapeutics, it is crucial for biomarker assays to compensate for inter-individual variabilities in measured values. Capturing multiple nuclear-originated and mitochondrial-originated tRFs using a dual primer qPCR blood-based assay may fulfill this demand. Specifically, our tRF-based score was designed to enable patient-tailored diagnosis, where the qPCR values determined for a certain patient will be compared to those of healthy volunteers or patients with PD of the same sex, ethnicity and genetic background, thus sparing the need for complex matching and normalization. Indeed, manual running of this dual qPCR test showed that the tRF-score segregated patients with PD from controls without the need for machine learning algorithms.
Notably, our study relied predominantly on samples from specific demographics, and, as such, the results might not be universally applicable to all populations. Thus, that the tRF-score was similar in prodromal African American patients and matched controls paralleled an identical trend in H&Y and UPDRS scores in this cohort, strengthening the notion that this score represents a true effect and calling for further studies of such patient-matched tests and validation in diverse patient cohorts.
Notably, too, blood RGTTCRA-tRF levels were effectively reduced under DBS stimulation, suggesting a linkage among tremor symptoms, electrical stimulus and the cellular localization and activities of these tRFs. Furthermore, yeast tRF research done by others43 and interaction predictions suggest that RGTTCRA-tRFs bind to the ribosome, potentially in an evolutionarily conserved translation-arresting manner. These findings are compatible with our FRET measurements and with the observation of ribosomal association and depolarization-triggered ribosomal dissociation of RGTTCRA-tRFs in cultured neuroblastoma cells. Moreover, as Ang may contribute to the synthesis of RGTTCRA-tRFs, our findings call for in-depth examination of the therapeutic potential of carefully adjusting its activity in the brains of patients with PD.
Mitochondrial death is a hallmark characteristic of many neurodegenerative diseases56,57 and was suggested as a blood biomarker in AD58,59. However, recent blood-based tests of mitochondrial dysfunction in PD14 reported that, in genetic cases, mitochondrial DNA damage was observed irrespective of a PD diagnosis14, highlighting the need to add other biomarkers to such tests. In this context, our combined nuclear/mitochondrial RGTTCRA-tRFs/MT-tRFs measurements enable segregating genetic PD patients from apparently healthy unaffected controls carrying the same PD-related mutations. An in-depth comprehensive analysis of larger patient cohorts will be needed to better determine the efficacy of our tRF-based measurement in comparison to other blood-based PD biomarkers.
The availability of an early, reliable and rapid blood test for PD diagnosis may potentially alleviate the burden of uncertainty from suffering individuals, assist clinicians’ care of those patients early in the disease process and open routes for developing and testing new therapeutics. To achieve these goals, an efficient diagnostic test should (1) provide accurate and disease-specific results, (2) avoid complex neurosurgical procedures, (3) entail minimal risks to the tested patients, (4) be easy to perform and (5) be inexpensive. Unlike CSF-based tests that can be risky, costly and dependent on complex molecular and biochemical procedures, qPCR-based blood tests are minimally invasive, do not depend on hospital care and offer quick procedures based on easily amplifiable signals. Our tRF-based blood measurement proved efficient in separating prodromal PD patients from controls by a simple dual qPCR assay, suggesting that it may equip clinicians with an informative diagnostic tool based on a single individual blood test, alone or in combination with other non-invasive measurements. The reliability, sensitivity, speed and low cost of our assay may all indicate its value for enabling an improved universal, pre-symptomatic PD diagnosis that can improve detection rates and offer beneficial clinical approaches to patients with PD at diverse stages of their disease.
Methods
Ethics statement
Ethics approval for this study was confirmed by the Hebrew University’s committee for research involving human-derived materials. Blood samples of the JLM cohort were collected at the Shaare Zedek Medical Center in Jerusalem under Helsinki approval number SZMC-0029-20.
Study population
This study consists of human brain (n = 8) and blood (n = 37) samples obtained from the NBB and of human brain samples (n = 23) obtained from the NIH NeuroBioBank, each under the relevant institution’s ethics and guidelines. Blood samples were further collected at the Shaare Zedek Medical Center (n = 30) under Helsinki approval number SZMC-0029-20. Other human data were downloaded from the PPMI (blood; n = 475) and the NIH (CSF; n = 152) and from GSE23676 (blood; n = 20), all under relevant ethics approval. Further details can be found in Extended Data Fig. 1 and in Supplementary Tables 1a, 3, 5–9 and 11–13. Written informed consent was obtained for all human samples included in this study.
Small RNA-seq of human SN samples
Brain tissues were obtained from the NBB as described in Hanan et al.4. RNA from eight male PD patient samples (see Supplementary Table 3 for patient information) was extracted using an miRNeasy Mini Kit (Qiagen, 217004) according to the manufacturer’s protocol, followed by RNA concentration determination (NanoDrop 2000; Thermo Fisher Scientific), standard gel electrophoresis for quality assessment and RIN determination (Bioanalyzer 6000; Agilent). Libraries were generated from 1,200 ng of RNA (NEBNext Multiplex Small RNA Library Prep Set for Illumina; New England Biolabs, E7560S), and the small RNA fraction was sequenced on a NextSeq 500 system (Illumina) at the Center for Genomic Technologies, Hebrew University of Jerusalem. Data were deposited in the Gene Expression Omnibus (GEO) (GSE256334). TH and DAT levels for these samples are taken from previously published data4.
Small RNA-seq of PM blood samples
Whole blood samples were obtained from the NBB (see Supplementary Table 5 for patient information). RNA was extracted using a NucleoSpin RNA Blood Mini Kit for RNA from blood (Machery-Nagel, 740200) according to the manufacturer’s protocol, followed by RNA concentration and quality determination as above. Libraries were generated from 100 ng of total RNA (NEBNext Multiplex Small RNA Library Prep Set for Illumina, as above), and the small RNA fraction was sequenced on a NextSeq 2000 system (Illumina) at the Center for Genomic Technologies, Hebrew University of Jerusalem. Data were deposited in GSE256334 as above.
Alignment of tRFs
FASTQ files (of both in-house sequencing and of publicly available data) were checked for quality using FastQC60, and adaptors (if present) were removed using FLEXBAR61 according to the pipeline manual. Adaptor-less output FASTQ files were then aligned to tRFs using MINTmap (version 1.0)62. See Extended Data Fig. 1 for elaboration.
Analysis of the CSF (ventricular) dataset
The CSF database phs000727 (downloaded from the National Center for Biotechnology Information (NCBI) database of Genotypes and Phenotypes (dbGaP)) included 66 patients with PD (20 removed as detailed below), 67 patients with AD (14 removed) and 69 healthy controls (16 removed). Before differential expression analysis, a principal component analysis was run based on tRF expression data, and samples that did not cluster were removed from the analysis. Those included six patients with PD, four patients with AD and six controls. Another five controls were removed because they had moderate SN depigmentation compared to no or mild depigmentation in other controls, and two additional controls were removed due to a combination of high Aβ plaque density (frequent) and high Tau tangle score (above 10). Likewise, 13 patients with PD were removed due to AD-like Braak, plaques or tangles scores. Seven patients with AD were removed due to severe SN depigmentation. Lastly, one control and one patient with AD were removed due to age younger than 60 years, and two controls, two patients with AD and one patient with PD were excluded due to exceptionally low tRF coverage (these patients had 21–1,627 counts, whereas the next lowest coverage patient had 11,745 counts). All eliminations were done before performing any analysis. See Supplementary Table 1a for general data of the CSF dataset and Supplementary Table 1b for a list of excluded samples. To further prevent any potential bias, differential expression analysis accounted for patient age, sex and PM interval.
Analysis of the PPMI dataset
Small RNA-seq was obtained in February 2021 from the PPMI database (https://www.ppmi-info.org/access-dataspecimens/download-data), RRID: SCR 006431. Updated participant status was re-downloaded in October 2023. For up-to-date information on the study, see https://www.ppmi-info.org/. Only samples with RIN ≥ 6 were analyzed, and we additionally filtered out samples of participants who were disqualified by the PPMI or who had undetermined sex63. See Supplementary Tables 6–8 and 11 for elaboration.
Data preparation and feature selection
To test our hypothesis that tRFs can constitute a patient-specific fingerprint, we used all the samples of PPMI patients having tRF counts from four different timepoints (n = 318). To focus on the most informative features, we selected the top 0.5% of enriched tRFs, resulting in a set of 253 tRFs for analysis. The data were split into training and test sets, with the test set comprising the last available sample from each patient, to evaluate the model’s performance across timepoints. Leveraging multiple timepoints per patient aimed to test whether blood-quantified tRFs might serve as a unique, patient-specific fingerprint and determine if the tRF values from a patient’s first three timepoints could accurately identify which of the remaining samples belonged to that patient. We implemented a MultiClassModel class to train and evaluate multiple machine learning classifiers, including XGBoost, HistGradientBoostingClassifier, RandomForestClassifier, Support Vector Classifiers (linear and nonlinear), K-Nearest Neighbors, Gaussian Naive Bayes and OneVsRest (OvR) Logistic Regression. The OvR approach, also called One-vs-All, extends binary classifiers such as logistic regression to handle multiclass problems by training a separate binary classifier for each class. In this setup, the class of interest is treated as the positive label, whereas all other classes are grouped as negative. During prediction, the classifier with the highest probability score is selected as the final output. OvR is computationally efficient and straightforward, although it may not capture interactions between classes as comprehensively as multinomial logistic regression or One-vs-One approaches. As supported by Rifkin and Klautau64, OvR can achieve accuracy similar to more complex methods, especially when binary classifiers are well tuned, making it a robust and widely used solution in practice. All features were standardized using StandardScaler before model training. The Matthews correlation coefficient (MCC) was chosen as the primary evaluation metric due to its robustness in assessing both binary and multiclass classifications. After identifying the best-performing classifier, we conducted hyperparameter tuning using GridSearchCV with predefined parameter grids, optimizing for the MCC score. To validate the assumption that the model’s performance was not due to chance, we compared the best-tuned classifier against a dummy model trained on shuffled labels. This comparison served to confirm the presence of a genuine, individual-specific signal in the tRF profiles.
For prodromal (pre-symptomatic) patient analysis, we subdivided, from the above-described PPMI data, prodromal and control patients with known genetic background (idiopathic or GBA+), known ethnicity, without prescribed PD medications and older than 58 years. Furthermore, patients with psychiatric anxiety, muscle inflammation, urinary bladder impairments and diabetes were excluded (due to underrepresentation in either the prodromal or the control group). To examine the earliest possible stage, we only selected patients with sequencing data from timepoints ‘BL’, ‘V02’ or ‘V04’ (corresponding to initial diagnosis, 6 months later and 12 months later). For each of these patients, we used the earliest timepoint available for the discovery sample (60 prodromal patients and 110 controls) and the second-earliest timepoint for the holdout validation sample (33 prodromal patients and 33 controls). The single Asian control meeting the above criteria was excluded because there was no Asian prodromal patient (see Supplementary Table 7 for sample information and the ‘Statistics and reproducibility’ subsection for classification processes). Additionally, to prevent biological outliers, we used only idiopathic patients of White ethnicity (as none of the other ethnicities or genetic backgrounds presented in both the prodromal and control groups to enable proper matching). Of note, as only one of the control patients in the test data had clinical scores (the rest of the controls had neither UPDRS nor H&Y scores), we imputed the UPDRS and the H&Y scores of the control patients. For each patient, we calculated the mean value of all the timepoints of that patient for each of these measurements. Because these were needed only for the control patients, the imputed values were anyway close to zero in both of the measurements.
To produce ROC curves for patients with PD, we selected patients and controls with known genetic background and without prescribed PD medications. To reduce variation between the groups, we selected patients only from ‘Phase 1’ and the following ethnicities: ‘White’, ‘Black or African American’, ‘Hispanic or Latino’ or ‘Ashkenazi Jewish’. Furthermore, to analyze the early stages of diagnosed PD patients, we used only timepoint ‘BL’ and PD patients for whom PD duration was 0. See Supplementary Table 8 for patient information.
To analyze DBS effects, we used small RNA-seq from the 19 patients with PD who were treated with DBS, all from the ‘Phase 2’ group and having familial PD, 14 with LRRK2 mutations and five with GBA mutations. Levels of RGTTCRA-tRFs in the DBS-treated patients were compared to those in PD patients with the above mutations who did not undergo DBS surgery (n = 11) and to those in control patients carrying the same mutations (n = 9). Non-DBS PD patients and controls were selected from the latest timepoint (V8) to account for the length of time most DBS-treated PD patients have been ill. See Supplementary Table 11 for sample information.
PPMI—a public–private partnership—is funded by the Michael J. Fox Foundation for Parkinson’s Research and by funding partners, including 4D Pharma, AbbVie, AcureX, Allergan, Amathus Therapeutics, Aligning Science Across Parkinson’s, AskBio, Avid Radiopharmaceuticals, BIAL, BioArctic, Biogen, Biohaven, BioLegend, BlueRock Therapeutics, Bristol Myers Squibb, Calico Labs, Capsida Biotherapeutics, Celgene, Cerevel Therapeutics, Coave Therapeutics, DaCapo Brainscience, Denali, the Edmond J. Safra Foundation, Eli Lilly, Gain Therapeutics, GE Healthcare, Genentech, GlaxoSmithKline, Golub Capital, Handl Therapeutics, Insitro, Janssen Neuroscience, Jazz Pharmaceuticals, Lundbeck, Merck, Meso Scale Discovery, Mission Therapeutics, Neurocrine Biosciences, Neuropore, Pfizer, Piramal, Prevail Therapeutics, Roche, Sanofi, Servier, Sun Pharma Advanced Research Company, Takeda, Teva, UCB, Vanqua Bio, Verily, Voyager Therapeutics, the Weston Family Foundation and Yumanity Therapeutics.
JLM blood sample set
Blood from 10 patients with PD, 10 patients with head trauma and 10 controls was taken in Jerusalem under Helsinki approval number SZMC-0029-20. Each group consisted of males and females, and all three had a similar average age (PD, 72 ± 7.5; Trauma, 73 ± 12; Control, 68 ± 5.6). See Supplementary Table 9 for detailed patient information. Samples were collected in Tempus Blood RNA Tubes (Thermo Fisher Scientific, 4342792), and RNA was extracted using a Norgen Preserved Blood RNA Purification Kit I (Norgen Biotek, 43400). Concentration was determined (NanoDrop 2000; Thermo Fisher Scientific), and quality was assessed by standard agarose gel electrophoresis. cDNA synthesis (from 250 ng) and qPCR are described below.
NIH NeuroBioBank sample set
SN tissue samples (left hemisphere) were obtained from the NIH NeuroBioBank. All donors died of natural cause at age 71–79 years and were of ‘White’ ethnicity. For elaboration, see Supplementary Table 10. RNA was extracted using the miRNeasy Mini Kit as above, followed by RNA concentration determination and quality assessment as above. cDNA synthesis (from 500 ng) and qPCR are described below.
cDNA synthesis and qPCR
cDNA was prepared using a qScript microRNA cDNA Synthesis Kit (Quantabio, 95107). qPCRs were performed in 96-well (Bio-Rad, HSP9601) or 384-well (Bio-Rad, HSP3805) plates, using PerfeCTa SYBR Green FastMix Low ROX (Quantabio, 95074) and primers (Merck) as below, on a Bio-Rad CFX384 Touch Real-Time PCR System with CFX Maestro software (Bio-Rad, version 4.1.2433.1219). The qPCR was programmed for 30 s of annealing/extension at 50 °C due to the short primer length. ΔCq for each sample was calculated as ‘mean (RGTTCRA-tRF triplicate) − mean (MT-tRF triplicate)’. ΔΔCq was calculated by dividing each ΔCq value by the mean ΔCq of the control patients. Expression of each sample was calculated as 2ΔΔCq.
Forward primer sequences:
-
a.
RGTTCRA-tRF primer for blood samples: GGTCCCTGGTTCAA
-
b.
RGTTCRA-tRF primer for brain samples: CGGGTTCGATTCCC
-
c.
MT-tRF primer for blood and brain samples: TAACTTAGCATTAACCTTTTAA
qPCR amplicon sequencing
The products of two qPCR amplicons (PD1 and T7) were sequenced as follows. Small RNA libraries were prepared from 500 ng of cDNA (blunting with NEB-E1201, followed by poly(A) and library preparation with KAPA (KK8420) mRNA-seq Kit) and sequenced using the NextSeq 2000 system (Illumina) as described above. FASTQ files were aligned to tRFs as described above.
Soreq DBS long-RNA array
Sequencing data of blood taken from six controls and seven patients with PD (all males) were downloaded from GEO dataset GSE23676. Blood from patients with PD was drawn before DBS surgery and several months after surgery, upon optimal stimulation (Supplementary Table 12). Affymetrix probes of Ang were found using biomaRt65, and all the Ang probes were summed to yield the expression of the gene.
Binding energy of tRFs to the ribosome-essential tRF and rRNAs
To assess the affinity of the CSF-expressed tRFs (1,017 tRFs in the CSF dataset that were expressed above a median of 10 CPM) to the Ribo-essential tRF44 and to rRNAs66, we used the RNAup web server45. We checked each of the expressed tRFs against the sequences of the Ribo-essential tRFs and against the open regions in the rRNAs that we found to contain reverse complement sequences to the RGTTCRA sequence (see sequences below), and we retrieved the total energy for each tRF (total energy equals the sum of melting energies of each of the sequences minus their binding energy to each other):
-
a.
Ribo-essential tRF: TCGAATCCCACTCCTGACACCA
-
b.
18S RGTTCRA reverse complement open region: AGGGGCGAAAGACTAATCGAACCAUTCTA
-
c.
28S RGTTCRA reverse complement open region: TAACCCGTTGAACCCCAT
Cell culture
The human-derived neuroblastoma cell line LAN-5 was cultured under standard conditions (37 °C, 5% CO2) in RPMI (Merck, R0883) supplemented with FCS (10% final concentration; Sartorius, 04-127), L-glutamine (2 mM final concentration; Sartorius, 03-020) and penicillin–streptomycin–amphotericin (100 U ml−1, 0.1 mg ml−1, 0.25 µg ml−1, final concentrations, respectively; Sartorius, 03-033). Cells were mycoplasma free (MycoBlue Mycoplasma Detection Kit; Vazyme, D101) and were used between passages 5 and 8.
Immunofluorescence and in situ hybridization
Cells were plated on glass coverslips in 24-well plates at 30,000 cells per well. Six days later, cells were fixed with 4% paraformaldehyde and permeabilized with methanol at −20 °C. Blocking solution (PBS with 5% normal donkey serum) was added to the wells for 1 h at room temperature, followed by 1-h incubation with primary antibody against human RPL24 (Proteintech, 17082-1-AP; rabbit polyclonal, 1:1,000) and secondary antibody labeled with Alexa Fluor 568 (Thermo Fisher Scientific, A10042; donkey anti-rabbit, 1:1,000) for 2 h. Cells were then washed with FISH buffer (1:1 of 2× SCC and 50% formamide and containing 50 μg ml−1 heparin), followed by overnight incubation at 37 °C with Affinity Plus (Integrated DNA Technologies) in situ probe, at a concentration of 200 nM. The probes were 5′-labeled with Alexa Fluor 488 and contained locked nucleic acids at the bases marked ‘+’:
-
a.
Probe sequence: ACT + CGAA + CCCA + CAAC + CTT
FRET-FLIM with PIE
To assess interactions with donor–acceptor proximity of less than 10 nm (refs. 48,67,68,69) between RGTTCRA-tRFs and ribosomes, we used FRET-FLIM with PIE70,71,72 (Extended Data Fig. 5). This allowed collecting fluorescence photon absolute detection times and detection times relative to moments of excitation for donor fluorescence after donor excitation, for acceptor fluorescence after donor excitation and FRET and for acceptor fluorescence after acceptor direct excitation. This was achieved with a confocal-based setup (ISS) assembled on top of a modified Olympus IX71 inverted microscope, as previously reported73,74. Donor and acceptor PIE was achieved by 488-nm and 532-nm picosecond pulsed lasers (λ = 488 nm, pulse width 80 ps full width at half maximum (FWHM), λ = 532 nm, pulse of 100 ps FWHM, rate; QuixX 488-60 PS, Omicron-Laserage, and FL-532-PICO, CNI), operating at 20-MHz repetition rate and interleaved by 25 ns. Of note, levels of donor fluorescence leakage into the acceptor fluorescence detection channel were negligible47. The laser beams passed through a single-mode polarization maintaining optical fiber (P1-405BPM-FC-Custom, specifications similar to those of PM-S405-XP, Thorlabs), a collimating lens (AC080-016-A-ML, Thorlabs), a halfwave plate (WPMP2-20(OD)-BB 550 nm, Karl Lambrecht Corp.) and a linear polarizer (DPM-100-VIS, Meadowlark Optics). Laser beams for acquiring images in laser scanning mode were reflected through galvo-scanning mirrors (6215H XY, Novanta Corp.) and scan lens (30 mm diameter × 50 mm focal length (FL), VIS-NIR Coated, Achromatic Lens, Edmund Optics) using a dichroic beam splitter at 405 nm, 488 nm, 532 nm and 640 nm (ZT405/488/532/640rpc-XT, Chroma). A ×100/1.45 numerical aperture (NA) oil objective (UPLSAPO100XO ×100 / 1.45 / oil / ∞ / 0.17/ OFN 26.5, Olympus) focused the light onto a small effective excitation volume. Scattered light was imaged on a CMOS camera (ThorCam, Thorlabs) using Airy ring pattern visualization. Fluorescence was collected through the same objective and focused with an achromatic lens (25 mm diameter × 100 mm FL, VIS-NIR Coated, Edmund Optics) onto a 100-μm-diameter pinhole and re-collimated with another achromatic lens (f = 100 mm; AC254-060-A, Thorlabs). Fluorescence was then split into two detection channels, 510/20 for AF488 and 698/70 for AF568, using a dichroic mirror (555-nm cutoff, FF555-Di03-25 × 36, Semrock) followed by bandpass filter 510/20 nm for wavelength <555 nm and 698/70 nm for wavelength >555 nm (FF03-510/20-25 and ff01-697/58-25, Semrock). Photons were detected using cooled hybrid photomultipliers (model R10467U-40, Hamamatsu) routed to a time-correlated single-photon counting module (SPC-150, Becker & Hickl) as its START signal (STOP signal was routed from the pulsed laser controller). For data acquisition, we used VistaVision software (version 4.2.095, 64-bit, ISS) in time-tagged time-resolved (TTTR) file format, and images were obtained using a laser scanning module in which a three-axis DAC module (ISS) synchronized data acquisition and control over the galvo-scanning mirrors. Images for FLIM measurements were obtained by tail-fitting fluorescence decays of each pixel in each photon stream (combination of excitation laser and detection channel), if it had at least 50 photons. FLIM images were acquired with 10-ms pixel dwell times in 15 × 15-μm2 image dimensions and 128 × 128-pixel resolutions.
Statistics and reproducibility
All multiple comparisons were FDR corrected, and all visualizations were done using ggplot2 (ref. 75) in R (version 4.0.3)76. All statistical tests were conducted two-sided unless reported otherwise. All of the box plots in this paper are defined so that the central line of the box plot represents the median; the lower and upper box bounds represent 0.25 and 0.75 quantiles, respectively; and lower and upper whisker bounds represent 0 and 1 quantiles, respectively. Differential analysis was carried out using edgeR77 (version 4.0.14) with patient age, sex and PM interval used as covariates in the design matrix of the edgeR analysis. Data collection and analysis were not performed blinded to the conditions of the experiments. No statistical methods were used to pre-determine sample sizes, but our sample sizes are compatible with those accepted in other publications in the field34,38,63. Due to the small number of samples in the SN data, we used the RNASeqPower pipeline78 (version 1.42.0) to estimate the power of the differentially expressed tRFs in these samples. We found the power of each of the differentially expressed tRFs to be higher than 0.87, with most of them (25th quantile and above) having a power higher than 0.9, which may improve the credibility of our findings in spite of the small number of samples.
Whenever the number of samples was smaller than 30, and the data did not distribute normally (P < 0.05 in a Shapiro test), a non-parametric test was run. Non-parametric tests were likewise used upon the need to compare median rather than mean values. All correlation tests conducted in this study (Extended Data Figs. 3c and 4e,f) are Spearman correlations, as we were interested in any type rather than linear correlation. Whenever comparing two groups to a third one, we used a Dunnett test (Figs. 1b,c and 3g,h and Extended Data Fig. 5a)
Binomial test was run on the differentially expressed tRFs in the CSF (Fig. 1b) and showed that they behave as a family and that most of the tRFs carrying the motif change in the same way. Upon comparing RGTTCRA-tRF and MT-tRF or RGTTCRA/MT-tRF ratio in blood of living and PM patients (Fig. 2f–h), we used the Mann–Whitney test (with FDR correcting for multiple comparisons) to compare the PD effect (PD versus Ctrl in each subgroup of a specific dataset and genetic background), and we employed a two-way ANOVA test to estimate the overall effect of PD while accounting for the dataset (that is, PM or alive) and the genetic background as covariates. The reason for using Mann–Whitney rather than a t-test for the first type of comparisons was that both the genetic controls and the PD idiopathic groups included low patient numbers (16 in each), and the data did not distribute normally (P < 0.0014, P < 0.003, P < 3 × 10−5, for PM RGTTCRA-tRF and MT-tRFs and Live genetic MT-tRFs, respectively; Shapiro test).
Matching analysis was carried out using MatchIt79 (version 4.6.0), with matched factors being sex, age and batch (‘Study’), distance = ‘glm’ and link = ‘probit’. For each of the ROC calculations (either prodromal and controls or PD and controls), we first used an ‘optimal’ matching in which each prodromal or PD patient was matched with a control (based on the above-mentioned factors). Next, the ratio between RGTTCRA-tRFs to MT-tRFs was normalized for each matched pair of prodromal and control or PD and control individuals, by dividing the pair’s scores by the pair’s mean. Lastly, we used a machine learning algorithm to calculate ROC (MLeval, version 0.3)80. In the feature selection process, a Mann–Whitney calculation was used to prevent outlier patients from skewing the results (for this reasoning, we focused on median rather than mean differences). The dimensions used for the ROC calculations included ethnicity and either UPDRS III and H&Y scores or the tRF normalized ratio (described above). The training control was performed with five cross-validations using both GBM and linear model (LM) machine learning algorithms (method = ‘cv‘, summaryFunction = twoClassSummary, number = 5 and classProbs and savePredictions = T). ROC curves were plotted using the pROC package81 (version 1.18.5).
In training the model on randomly mixed labels, we received an AUC of 0.43, which reflects systematic misclassification rather than predictive power, as the model picked up on random patterns in the data. This result illustrates the random distribution expected in a sham model setup, with a marginal error. We further ran iterations of a ‘full’ matching in which patients were divided into clusters, each containing at least one prodromal or PD patient and one control, and the same pipeline as described above for the ‘optimal’ matching was used.
To compute the ability of the tRF-based and the clinical-based algorithms to classify patients (Extended Data Fig. 4g), we ran chi-square, first using the percentage of patients predicted by random distribution and then using the percentage of patients predicted by the clinical-based algorithm as the reference probability. This created a confusion matrix of two columns (predicted to be prodromal, predicted to be control) and two rows (is indeed prodromal, is indeed control). Comparing the matrix of the tRF-based algorithm to the reference clinical-based matrix enabled proving that the tRF-based algorithm predicted a higher number of true prodromal patients to be prodromal.
Checking random motifs for the ability to segregate prodromal from control patients
First, we extracted all options of 14-nt-long sequences found in the PPMI-expressed tRFs. For example, for a tRF with the length of 30 nt, we regarded each of the sequences produced from nucleotides 1–14, 2–15, 3–16… 17–30 as a potential motif. We than created 10,000 couples of randomly selected motifs and, for each couple, ran the same process as described above (namely, for each patient, we summed the counts for each of these motifs and calculated the ratio between the sums). We then normalized the ratio for each pair of optimally matched prodromal and control patients by dividing both by the mean of the pair’s ratio. We ran GMB analysis and extracted the yielded AUC.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All RNA sequencing data created for this study are available in the GEO under GSE256334. Data produced by others and used for this study are available at the NIH under phs000727, in the PPMI consortium and in the GEO under GSE130764, GSE23676 and GSE155727. Metadata and column data for the used datasets are available in the supplementary tables. Any other data reported in this paper are available from the lead contact upon reasonable request.
Code availability
The codes used for these data are available at https://github.com/NimrodMd/PD_motif_codes.git.
References
Aarsland, D. et al. Parkinson disease-associated cognitive impairment. Nat. Rev. Dis. Primers 7, 47 (2021).
Parkinson’s disease economic burden on patients, families and the federal government is $52 billion, doubling previous estimates. https://www.michaeljfox.org/publication/parkinsons-disease-economic-burden-patients-families-and-federal-government-52-billion (2019).
Trist, B. G., Hare, D. J. & Double, K. L. Oxidative stress in the aging substantia nigra and the etiology of Parkinson’s disease. Aging Cell 18, e13031 (2019).
Hanan, M. et al. A Parkinson’s disease CircRNAs Resource reveals a link between circSLC8A1 and oxidative stress. EMBO Mol. Med. 12, e11942 (2020).
Stykel, M. G. & Ryan, S. D. Nitrosative stress in Parkinson’s disease. NPJ Parkinsons Dis. 8, 104 (2022)
Danielson, S. R. & Andersen, J. K. Oxidative and nitrative protein modifications in Parkinson’s disease. Free Radic. Biol. Med. 44, 1787–1794 (2008).
Masaldan, S., Callegari, S. & Dewson, G. Therapeutic targeting of mitophagy in Parkinson’s disease. Biochem. Soc. Trans. 50, 783–797 (2022).
Lane, J. D. et al. Autophagy impairment in Parkinson’s disease. Essays Biochem. 61, 711–720 (2017).
Zhang, Y. F. & Cragg, S. J. Revisiting dopamine-acetylcholine imbalance in Parkinson’s disease: glutamate co-transmission as an exciting partner in crime. Neuron 109, 1070–1071 (2021).
Tolosa, E., Garrido, A., Scholz, S. W. & Poewe, W. Challenges in the diagnosis of Parkinson’s disease. Lancet Neurol. 20, 385–397 (2021).
Eusebi, P. et al. Diagnostic utility of cerebrospinal fluid α-synuclein in Parkinson’s disease: a systematic review and meta-analysis. Mov. Disord. 32, 1389–1400 (2017).
Poggiolini, I. et al. Diagnostic value of cerebrospinal fluid alpha-synuclein seed quantification in synucleinopathies. Brain 145, 584–595 (2022).
Grossauer, A. et al. α-Synuclein seed amplification assays in the diagnosis of synucleinopathies using cerebrospinal fluid—a systematic review and meta-analysis. Mov. Disord. Clin. Pract. 10, 737–747 (2023).
Qi, R. et al. A blood-based marker of mitochondrial DNA damage in Parkinson’s disease. Sci. Transl. Med. 15, eabo1557 (2023).
Magalhães, P. & Lashuel, H. A. Opportunities and challenges of alpha-synuclein as a potential biomarker for Parkinson’s disease and other synucleinopathies. NPJ Parkinsons Dis. 8, 93 (2022).
Leek, J. T. svaseq: removing batch effects and other unwanted noise from sequencing data. Nucleic Acids Res. 42, e161 (2014).
McMahon, S. S., Lenive, O., Filippi, S. & Stumpf, M. P. H. Information processing by simple molecular motifs and susceptibility to noise. J. R. Soc. Interface 12, 0597 (2015).
Madrer, N. & Soreq, H. Cholino-ncRNAs modulate sex-specific- and age-related acetylcholine signals. FEBS Lett. 594, 2185–2198 (2020).
Guo, D. & Li, C. Stochastic and coherence resonance in feed-forward-loop neuronal network motifs. Phys. Rev. E 79, 051921 (2009).
Shen-Orr, S. S., Milo, R., Mangan, S. & Alon, U. Network motifs in the transcriptional regulation network of Escherichia coli. Nat. Genet. 31, 64–68 (2002).
Ross, C. J. et al. Uncovering deeply conserved motif combinations in rapidly evolving noncoding sequences. Genome Biol. 22, 29 (2021).
Gaetani, L. et al. CSF and blood biomarkers in neuroinflammatory and neurodegenerative diseases: implications for treatment. Trends Pharmacol. Sci. 41, 1023–1037 (2020).
Shreffler, J. & Huecker, M. R. Diagnostic Testing Accuracy: Sensitivity, Specificity, Predictive Values and Likelihood Ratios (StatPearls, 2023).
Grigoriev, A. & Karaiskos, S. Dynamics of tRNA fragments and their targets in aging mammalian brain. F1000Res. 5, 2758 (2016).
Kumar, P., Kuscu, C. & Dutta, A. Biogenesis and function of transfer RNA-related fragments (tRFs). Trends Biochem. Sci. 41, 679–689 (2016).
Ivanov, P., Emara, M. M., Villen, J., Gygi, S. P. & Anderson, P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell 43, 613–623 (2011).
Magee, R. & Rigoutsos, I. On the expanding roles of tRNA fragments in modulating cell behavior. Nucleic Acids Res. 48, 9433–9448 (2020).
Bose, A. & Beal, M. F. Mitochondrial dysfunction in Parkinson’s disease. J. Neurochem. 139, 216–231 (2016).
Toomey, C. E. et al. Mitochondrial dysfunction is a key pathological driver of early stage Parkinson’s. Acta Neuropathol. Commun. 10, 134 (2022).
Prehn, J. H. M. & Jirström, E. Angiogenin and tRNA fragments in Parkinson’s disease and neurodegeneration. Acta Pharmacol. Sin. 41, 442–446 (2020).
Sheng, J. & Xu, Z. Three decades of research on angiogenin: a review and perspective. Acta Biochim. Biophys. Sin. 48, 399–410 (2016).
Magee, R., Londin, E. & Rigoutsos, I. TRNA-derived fragments as sex-dependent circulating candidate biomarkers for Parkinson’s disease. Parkinsonism Relat. Disord. 65, 203–209 (2019).
Thomas, S. P., Hoang, T. T., Ressler, V. T. & Raines, R. T. Human angiogenin is a potent cytotoxin in the absence of ribonuclease inhibitor. RNA 24, 1018–1027 (2018).
Winek, K. et al. Transfer RNA fragments replace microRNA regulators of the cholinergic poststroke immune blockade. Proc. Natl Acad. Sci. USA 117, 32606–32616 (2020).
Hogg, M. C. et al. 5′ValCAC tRNA fragment generated as part of a protective angiogenin response provides prognostic value in amyotrophic lateral sclerosis. Brain Commun. 2, fcaa138 (2020).
Hogg, M. C. et al. Elevation of plasma tRNA fragments precedes seizures in human epilepsy. J. Clin. Invest. 129, 2946–2951 (2019).
Paldor, I. et al. Cerebrospinal fluid and blood profiles of transfer RNA fragments show age, sex, and Parkinson’s disease-related changes. J. Neurochem. 164, 671–683 (2023).
Burgos, K. et al. Profiles of extracellular miRNA in cerebrospinal fluid and serum from patients with Alzheimer’s and Parkinson’s diseases correlate with disease status and features of pathology. PLoS ONE 9, e94839 (2014).
Bartel, D. P. Metazoan microRNAs. Cell 173, 20–51 (2018).
Bailey, T. L. & Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36 (1994).
Su, Z., Kuscu, C., Malik, A., Shibata, E. & Dutta, A. Angiogenin generates specific stress-induced tRNA halves and is not involved in tRF-3-mediated gene silencing. J. Biol. Chem. 294, 16930–16941 (2019).
Ferreira, P. G. et al. The effects of death and post-mortem cold ischemia on human tissue transcriptomes. Nat. Commun. 9, 490 (2018).
Bąkowska-Żywicka, K., Kasprzyk, M. & Twardowski, T. tRNA-derived short RNAs bind to Saccharomyces cerevisiae ribosomes in a stress-dependent manner and inhibit protein synthesis in vitro. FEMS Yeast Res. 16, fow077 (2016).
Kim, H. K. et al. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature 552, 57–62 (2017).
Gruber, A. R., Lorenz, R., Bernhart, S. H., Neuböck, R. & Hofacker, I. L. The Vienna RNA Websuite. Nucleic Acids Res. 36, W70–W74 (2008).
Darnell, A. M., Subramaniam, A. R. & O’Shea, E. K. Translational control through differential ribosome pausing during amino acid limitation in mammalian cells. Mol. Cell 71, 229–243 (2018).
Razvag, Y., Drori, P., Klemfner, S., Meshorer, E. & Lerner, E. FRETsael: nanometer localization of biomolecular interactions using fluorescence lifetime imaging. Biophys. J. https://doi.org/10.1016/j.bpj.2025.02.017 (2025).
Lerner, E. et al. Toward dynamic structural biology: two decades of single-molecule Förster resonance energy transfer. Science 359, eaan1133 (2018).
Lozano, A. M. et al. Deep brain stimulation: current challenges and future directions. Nat. Rev. Neurol. 15, 148–160 (2019).
Ashkan, K., Rogers, P., Bergman, H. & Ughratdar, I. Insights into the mechanisms of deep brain stimulation. Nat. Rev. Neurol. 13, 548–554 (2017).
Goldwurm, S. et al. Evaluation of LRRK2 G2019S penetrance. Neurology 68, 1141–1143 (2007).
Soreq, L. et al. Deep brain stimulation induces rapidly reversible transcript changes in Parkinson’s leucocytes. J. Cell. Mol. Med. 16, 1496–1507 (2012).
Kiltschewskij, D. J. & Cairns, M. J. Transcriptome-wide analysis of interplay between mRNA stability, translation and small RNAs in response to neuronal membrane depolarization. Int. J. Mol. Sci. 21, 7086 (2020).
Leyton, M. et al. Effects on mood of acute phenylalanine/tyrosine depletion in healthy women. Neuropsychopharmacology 22, 52–63 (2000).
Lyons, J. et al. Blood glutathione synthesis rates in healthy adults receiving a sulfur amino acid-free diet. Proc. Natl Acad. Sci. USA 97, 5071–5076 (2000).
Wang, W., Zhao, F., Ma, X., Perry, G. & Zhu, X. Mitochondria dysfunction in the pathogenesis of Alzheimer’s disease: recent advances. Mol. Neurodegener. 15, 30 (2020).
Knott, A. B., Perkins, G., Schwarzenbacher, R. & Bossy-Wetzel, E. Mitochondrial fragmentation in neurodegeneration. Nat. Rev. Neurosci. 9, 505–518 (2008.
Lee, J. W., Park, K. D., Im, J. A., Kim, M. Y. & Lee, D. C. Mitochondrial DNA copy number in peripheral blood is associated with cognitive function in apparently healthy elderly women. Clin. Chim. Acta 411, 592–596 (2010).
Perez Ortiz, J. M. & Swerdlow, R. H. Mitochondrial dysfunction in Alzheimer’s disease: role in pathogenesis and novel therapeutic opportunities. Br. J. Pharmacol. 176, 3489–3507 (2019).
Andrews, S. s-andrews/FastQC: a quality control analysis tool for high throughput sequencing data. Github https://github.com/s-andrews/FastQC (2023).
Dodt, M., Roehr, J. T., Ahmed, R. & Dieterich, C. FLEXBAR—flexible barcode and adapter processing for next-generation sequencing platforms. Biology 1, 895–905 (2012).
Loher, P., Telonis, A. G. & Rigoutsos, I. MINTmap: fast and exhaustive profiling of nuclear and mitochondrial tRNA fragments from short RNA-seq data. Sci. Rep. 7, 41184 (2017).
Craig, D. W. et al. RNA sequencing of whole blood reveals early alterations in immune cells and gene expression in Parkinson’s disease. Nat. Aging 1, 734–747 (2021).
Rifkin, R. & Klautau, A. In defense of one-vs-all classification. J. Mach. Learn. Res. 5, 101–141 (2004).
Durinck, S., Spellman, P. T., Birney, E. & Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 4, 1184–1191 (2009).
Sweeney, B. A. et al. Exploring non-coding RNAs in RNAcentral. Curr. Protoc. Bioinformatics 71, e104 (2020).
van der Meer, B. W. in FRET – Förster Resonance Energy Transfer (eds Medintz, I. & Hildebrandt, N.) 23–62 (Wiley, 2013).
Lakowicz, J. R. Principles of Fluorescence Spectroscopy 3rd edn (Springer, 2006).
Stryer, L. & Haugland, R. P. Energy transfer: a spectroscopic ruler. Proc. Natl Acad. Sci. USA 58, 719–726 (1967).
Laptenok, S. P. et al. Global analysis of Förster resonance energy transfer in live cells measured by fluorescence lifetime imaging microscopy exploiting the rise time of acceptor fluorescence. Phys. Chem. Chem. Phys. 12, 7593–7602 (2010).
Reissaus, C. A. et al. PIE-FLIM measurements of two different FRET-based biosensor activities in the same living cells. Biophys. J. 118, 1820–1829 (2020).
Müller, B. K., Zaychikov, E., Bräuchle, C. & Lamb, D. C. Pulsed interleaved excitation. Biophys. J. 89, 3508–3522 (2005).
Joron, K. et al. Fluorescent protein lifetimes report densities and phases of nuclear condensates during embryonic stem-cell differentiation. Nat. Commun. 14, 4885 (2023).
Chen, J. et al. The structural heterogeneity of α-synuclein is governed by several distinct subpopulations with interconversion times slower than milliseconds. Structure 29, 1048–1064 (2021).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer, 2016).
R Core Team R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2020); https://www.r-project.org/
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Hart, S. N., Therneau, T. M., Zhang, Y., Poland, G. A. & Kocher, J. P. Calculating sample size estimates for RNA sequencing data. J. Comput. Biol. 20, 970–978 (2013).
Ho, D. E., Imai, K., King, G. & Stuart, E. A. MatchIt: nonparametric preprocessing for parametric causal inference. J. Stat. Softw. 42, 1–28 (2011).
John, C. R. MLeval: machine learning model evaluation. R version 0.3 https://cran.r-project.org/web/packages/MLeval/index.html (2020).
Robin, X. et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatiocs 12, 77 (2011).
Acknowledgements
We acknowledge donations of blood, postmortem brain tissues and clinical data by numerous patients as well as support for this study by the Israel Science Foundation, grant numbers 1016/18, 3213/19, 1770/20 and 835/23; Michael J. Fox Foundation for Parkinsonʼs Research grant number 11183; and the ERA-NET NEURON_RV-025 B3phrenia project to H.S. We further acknowledge the Edmond and Lily Safra Center for Brain Sciences and national postdoctoral fellowships to T.Z., the support of Ken Stein and Keter Holdings and PhD fellowship support from the Leopold, Norman and Sara Yisraeli Memorial Fund and the Sephardic Home for the Aged to N.M. and S.V.-T. E.L. and P.D. were supported by Israel Science Foundation grant number 556/22. Patient brain tissues were obtained from the Netherlands Brain Bank and from the NIH NeuroBioBank. Blood samples and data were gratefully received from the Netherlands Brain Bank, from the Shaare Zedek Medical Center under joint grant support to I.P. and H.S. and from the Parkinson’s Progression Markers Initiative (PPMI) database (https://www.ppmi-info.org/access-data-specimens/download-data), RRID: SCR 006431. For up-to-date information on the study, see https://www.ppmi-info.org/ (see supplementary acknowledgement for elaboration). This paper further includes data under US provisional patent application number 63/531,628, titled ‘Methods for detection, qualification, or monitoring of Parkinson’s disease’, and application number 63/557,661, titled ‘Angiogenin-reducing agents for treating Parkinson’s disease’. We thank S. Shifman for wise advice, H. Bergman for introduction to the challenges of DBS and N. Swissa for patient blood samples. This work is dedicated to the memory of Mrs. Lily Safra, a great supporter of brain research.
Author information
Authors and Affiliations
Contributions
N.M. initiated the research, ran the analysis, created the figures and wrote the paper. S.V.-T. extracted RNA and prepared the NBB blood data for sequencing, coordinated the interaction with the PPMI consortium, reviewed the paper and advised on graphical matters. T.Z. extracted RNA from the NBB SN samples, revised the text and advised on graphical matters. Y.T. helped with qPCR and imaging processes and revised the text and the methods. E.R.B. extracted RNA from the JLM blood samples and thoroughly revised the main text and the methods. P.D. conducted the FRET measurements. N.S. interviewed patients and collected blood samples for the JLM cohort. D.S.G. ran the qPCR tests for the SN and JLM cohorts and revised the main text. E.L. orchestrated FRET measurements and revised the text. E.S. advised the statistical and computational analysis and conducted the fingerprint analysis. I.P. advised on clinical matters and revised the text. H.S. orchestrated the research, supplied the means and facilities for running this research and revised the text, the methods and the figures.
Corresponding authors
Ethics declarations
Competing interests
H.S., N.M., E.L. and S.V.-T. have patents listed on the finding of the PD motif sequence: application 63/531,628, titled ‘Methods for detection, qualification, or monitoring of Parkinson’s disease’, and application number 63/557,661, titled ‘Angiogenin-reducing agents for treating Parkinson’s disease’. H.S. serves as the senior academic advisor to the Azrieli Fellows Program in Israel. The other authors declare no competing interests.
Peer review
Peer review information
Nature Aging thanks Joanne Trinh, Panpan Zhang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Technical representation of the datasets and methods.
a. The different datasets used in this work with total number of samples in each. All datasets were subjected to files quality check (using FASTQC), adaptor removal (FLEXBAR) and alignment to tRFs (MINTmap). b. DE analysis using EdgeR was applied on CSF (PD vs. Ctrl) and SN (high vs. low Lewy body score) samples. c. The consensus motif sequence identified by the MEME tool. Letters heights: Nucleotides conservation among upregulated N-tRFs. d. PPMI samples were further divided to subgroups according to the relevant comparison, with prodromal PD patients (n = 189) divided to 60 + 60 optimally matched patients and controls, 33 + 33 optimally matched test data, and 60 + 110 fully matched patients and controls. e, f. Calculated sum (E) and percentage (F) for RGTTCRA-tRFs and MT-tRFs (and their ratio – RGTTCRA-tRFs/MT-tRFs) from all blood samples. g. Normalized calculated PD/MT-tRFs ratio for the two prodromal groups. h. The optimally matched patients served as a training dataset (GBM, k = 5 folds). i. Naïve test data and Fully matched patients served as a testing dataset. j. Blood samples collected in Jerusalem and SN samples obtained from the NIH NeuroBioBank served to test qPCR segregation of PD from controls using the primers designed based on the PPMI prodromal data. k. Long-RNA Affymetrix data (GSE23676) served to map Angiogenin levels in 7 PD patients before and after DBS neurosurgery.
Extended Data Fig. 2 MTtRFs decline, NtRFs and RGTTCRA-tRFs elevate in both Male and Female PD patients vs. controls.
a. Density plots of post-mortem interval in female and male samples (left, right) and patients’ age (upper and lower panel). Note that PD, but not AD appears to shorten patients’ life span. b, c. Volcano plots for male (A) and Female (B) CSF samples. Each dot is a tRF. X: log2(Fold Change) of PD/Ctrl levels. Y: -log10(FDR adjusted p value). Horizontal and vertical lines: FDR < 0.05; log2(FC) > 1 or <-1. Left: NtRFs, right: MTtRFs. Dot colors: tRFs’ length. d. Scheme of tRNA breakdown into tRF types by Angiogenin (Ang) or Dicer (DICR). e. Segregation of PD-modified tRFs (Fig. 1b) to subtypes as in A. Each dot is a tRF. X: log2(Fold Change) of PD/Ctrl levels. Y: -log10(FDR adjusted p value). Background colors: NtRFs (blue), MTtRFs (green). f, g. Volcano plots for male (E) and Female (F) CSF samples. PD CSF NtRFs segregated into RGTTCRA-carrying and lacking tRFs. Blue thick line: FDR = 0.05. Grey line: unadjusted p = 0.05. Each dot is a tRF. X: log2(Fold Change) of PD/Ctrl levels. Y: -log10(FDR adjusted p value). Dot colors: tRFs’ length. h. As in A for 8 PD SN samples.
Extended Data Fig. 3 Overexpression but not knockout of Ang selectively affect RGTTCRA-tRF levels which also correlate to Lewy bodies score in PD blood. MT-tRNA are reduced with PD duration.
a. KO of Ang (green) in U2SO cells with or without sodium arsenite (SA) exposure (purple and grey). Y axis: percentage of RGTTCRA-tRFs out of all expressed tRFs. b. As A. for MT-tRFs percentage. Both A and B were analyzed with a two-way ANOVA test that yielded no significant results. c. SN samples of healthy controls with (grey) or without (pink) healthy-reflecting levels of TH in their SN, compared to PD patients (blue) and SWEDD-like patients (orange) from a cohort of NBB Long-RNA-seq (n = 25). Y axis: Log10 of sum of all expressed MT-tRNAs. X axis: PD duration in years. r = 0.4, p < 0.05, Spearman correlation; PD vs. Ctrl p < 0.039 two-sided Mann-Whitney test. All boxplots in this figure are defined so that the central line of the boxplot represents the median, the lower and upper box bounds represent 0.25 and 0.75 quantiles and lower and upper whiskers bounds represent 0 and 1 quantiles.
Extended Data Fig. 4 RGTTCRA/MT-tRF score separates prodromal and controls even without normalization and UPDRS.III scores show similar, yet less significant, trends to RGTTCRA/MT-tRFs scores.
a. ROC curve of the GLM algorithm classifying prodromal and control patients, based on only tRFs (tRFs-score; orange), only clinical measurements (UPDRS section III plus Hoehn and Yahr; turquoise) or Null classification (tRFs + clinical measurements albeit with mixed labels; grey). b. Ratio between RGTTCRA-tRFs to MT-tRFs in Prodromal patients (orange; n = 60) and controls (grey; n = 129) separated by their ethnicity and genetic backgrounds. Prodromal vs. Ctrl p < 5 × 10−6, two-way ANOVA c. UPDRS.III (motor) score in optimally matched Prodromal patients (orange; n = 60) and controls (grey; n = 60) separated by their ethnicity and genetic backgrounds. d. UPDRS III scores (top) and tRFs-score (bottom) for control, prodromal and PD patients according to their ethnicities and genetic backgrounds. UPDRS: Prodromal vs. Ctrl p < 0.11, PD vs. Ctrl p < 1 × 10−8, PD vs. prodromal p < 1 × 10−8, two-way ANOVA. tRFs-score: Prodromal vs. Ctrl p < 1 × 10−7, PD vs. Ctrl p < 1 × 10−4, PD vs. prodromal p < 0.043, two-way ANOVA. e. Correlation between total UPDRS scores (Y) and tRFs-score (X axis of E) in optimally matched prodromal and control patients (n = 60 of each). r = 0.27, p < 0.01; Spearman correlation, FDR correction. f. As in E for motoric (section III) UPDRS and for normalized RGTTCRA-tRF and MT-tRF levels (purple and green, X axis of F) separately. I: r = 0.19, p < 0.06; II: r = 0.245, p < 0.0163; III: r = 0.318, p < 0.004; Spearman correlation, FDR correction. g. GBM algorithm trained in Fig. 3b applied onto fully-matched prodromal and control subjects (n = 60, n = 110). X axis: true classification. Y axis: prospects to be a prodromal PD patient based on the algorithm. tRFs: p < 5 × 10−14, UPDRS: p < 3 × 10−7, Chi-square test, FDR correction. h. Each dot represents a prodromal patient that was later diagnosed as PD (n = 21). X axis: UPDRS score (at basal level, not at diagnosis as PD), Y axis: tRFs-score. i. Amplicons of two qPCR products (mixture of all three triplicates) were sequenced and aligned to tRFs. >50% of the reads were mapped to different tRFs (each color represents a single tRF. Note that the most prominent tRF in sample PD1 is hardly expressed in T7 and vice versa. All boxplots in this figure are defined so that the central line of the boxplot represents the median, the lower and upper box bounds represent 0.25 and 0.75 quantiles and lower and upper whiskers bounds represent 0 and 1 quantiles.
Extended Data Fig. 5 Ribosome profiling and FRET imaging suggest direct interaction of RGTTCRA-tRFs with the ribosome.
a. Interaction of motif-carrying tRFs with ribosomes based on the GSE113751 dataset of short RNA-seq from ribosomes pulldown in HeLa (red), HCT116 (green) and HEK293T (blue) cells. Cells were either untreated or starved for Arg(inine) or Leu(cine) for 3 or 6 hours. Left Y axis: Points show RGTTCRA- tRF fractions among all tRFs. Right Y axis: total number of reads per sample. Horizontal dashed line: percent RGTTCRA-tRFs among all tRFs. b. Fluorescence intensity image of RGTTCRA-tRF (green), ribosomes (RPL24, red) and their colocalization (yellow). c. FRET-sensitized fluorescence of the acceptor indicates a non-negligible amount of donor-labeled RGTTCRA-tRF and acceptor-labeled antibody-tagged RPL24 ribosomal protein found at proximities <10 nm, and hence tRF and ribosomes interact. d, e. Fluorescence lifetime image of FRET-sensitized (C) or directly excited (D) acceptor-labeled ribosomes. f. The acceptor dye labeled by an antibody tagging the ribosomal protein RPL24 presents acceptor fluorescence decay of a region of interest (green & red arrows) after direct (red) and FRET-sensitized excitation, reflected as slower fluorescence decay of FRET-sensitized compared to direct excitation events. C, D, E: ROIs with tRFs-ribosome interactions (arrows).
Extended Data Fig. 6 Two hours depolarization impairs ribosome enrichment of RGTTCRA-containing, 3′-tRFs and i-tRFs in cultured neuroblastoma cells; but MT-tRFs do not present significant changes under DBS stimuli.
a. Blood MT-tRF levels in apparently healthy control carriers of PD-related mutations (Ctrl; grey; n = 9) and in PD patients, carriers of mutations in the LRRK2 and GBA genes (PD; blue; n = 30), with and without DBS (black and red lines; n = 11, n = 19). Y axis: mean blood MT-tRFs levels (white rhombuses). No significant changes, one-way ANOVA. b. GSE155727 dataset of ribosomal profiling and short-RNA-seq from SHSY cells. Counts per million of 3′-tRFs and i-tRFs lacking our motif (green) and carrying it (purple), from ribosome associated RNA-seq (left) or bulk short RNA-seq (right) in resting cells, or in cells right after or two hours post depolarization (2 h pDP). All boxplots in this figure are defined so that the central line of the boxplot represents the median, the lower and upper box bounds represent 0.25 and 0.75 quantiles and lower and upper whiskers bounds represent 0 and 1 quantiles.
Supplementary information
Supplementary Information
Supplementary information for Extended Data Fig. 2, Extended Data Fig. 3, ‘Blood tRF levels create a patient-specific fingerprint’, ‘Motif-carrying tRFs with ribosomes’ and Extended Data Fig. 6.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Madrer, N., Vaknine-Treidel, S., Zorbaz, T. et al. Pre-symptomatic Parkinson’s disease blood test quantifying repetitive sequence motifs in transfer RNA fragments. Nat Aging 5, 868–882 (2025). https://doi.org/10.1038/s43587-025-00851-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43587-025-00851-z