Abstract
Human adipose depots are functionally distinct. Yet, recent single-nucleus RNA sequencing (snRNA-seq) analyses largely uncovered overlapping or similar cell-type landscapes. We hypothesized that adipocyte subtypes, differentiation trajectories and/or intercellular communication patterns could illuminate this depot similarity–difference gap. For this, we performed snRNA-seq of human subcutaneous or visceral adipose tissues (five or ten samples, respectively). Of 27,665 adipocyte nuclei in both depots, most were ‘classical’, namely enriched in lipid metabolism pathways. However, we also observed ‘nonclassical’ adipocyte subtypes, enriched in immune-related, extracellular matrix deposition (fibrosis), vascularization or angiogenesis or ribosomal and mitochondrial processes. Pseudo-temporal analysis showed a developmental trajectory from adipose progenitor cells to classical adipocytes via nonclassical adipocytes, suggesting that the classical state stems from loss, rather than gain, of specialized functions. Last, intercellular communication routes were consistent with the different inflammatory tone of the two depots. Jointly, these findings provide a high-resolution view into the contribution of cellular composition, differentiation and intercellular communication patterns to human fat depot differences.
Similar content being viewed by others
Main
The past ~30 years have revolutionized our understanding of adipose tissues and their roles in human health and disease. All adipose tissues, at their various anatomical locations, were found to be highly secretory, engaging through peptides, lipids, nucleotides and exosomes in active autoparacrine and endocrine communication, through which they regulate fundamental processes, including energy metabolism, immune function, vascular tone and reproduction.
Human subcutaneous adipose tissue (hSAT) and human visceral adipose tissue (hVAT) affect health differently, with hVAT accumulation being more detrimental in obesity1,2,3. Even in normal physiology, the two fat depots differ: hVAT is more lipolytic, possibly as a result of being more sympathetically innervated4; adipokine expression and secretion5 and immune cell composition differ, with hVAT exhibiting a more proinflammatory profile6. Collectively, these differences manifest in distinct whole-tissue transcriptional landscapes7, including distinct developmental paths of the two tissues8.
Recently, several adipose tissue single-cell resolution atlases have been reported9,10,11,12,13,14,15. Adipocytes, the size, fragility and lipid content of which render them unamenable to scRNA-seq, were included in recent atlases that utilized spatial and/or single-nucleus transcriptomics. These studies revealed adipocytes’ spatial arrangements and differential insulin responsiveness11, thermogenic adipocytes13, plasticity of mouse adipose tissue (AT) in response to diet-induced obesity14, adipogenic potential of distinct progenitor cells12, associations between specific adipose tissue cell types and metabolic states9,10 and commonalities or differences across species and dietary conditions9. Although depot differences have been noted, AT cell atlases mainly emphasized commonalities in cell composition between hSAT and hVAT, including in adipocyte subtypes. Hence, a major gap in our understanding remains: functionally and at the whole-tissue transcriptomics level, hSAT and hVAT are distinct. In contrast, at the single-cell level, particularly when focusing on adipocyte subtype landscape, the two fat depots overall seem quite similar.
In the present study, we utilized snRNA-seq to compare hSAT and hVAT adipocytes, creating depot-specific, adipocyte subtype atlases. We highlighted depot commonalities and differences in adipocyte subtype composition, differentiation trajectories and intercellular communication patterns, offering a new, high-resolution view into their functional roles.
Results
Human fat depot-specific adipocyte atlases
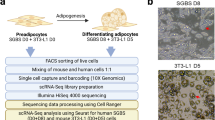
To generate hSAT and hVAT depot-specific human adipocyte atlases, we performed single-nucleus RNA sequencing (snRNA-seq) on five hSAT–hVAT paired samples and five additional samples of hVAT. Samples were donated by a relatively healthy cohort of men (20%) and women (80%) undergoing elective abdominal surgeries, with diverse body mass indices (BMIs; 23.8–44.1 kg m−2), ages (24.5–53.4 years) and backgrounds (Methods and Fig. 1a). A total of 37,879 hSAT nuclei and 83,731 hVAT nuclei passed quality control scrutiny (Methods and Supplementary Table 1) and underwent depot-separate clustering based on their nuclear RNA repertoires. Adipocytes were easily identified in both hSAT and hVAT by expressing adiponectin (ADIPOQ) and additional adipocyte markers AQP7, LPL and PLIN1 (Fig. 1b,c). Moreover, genes overexpressed in adipocyte clusters (>2-fold versus all other depot’s nuclei, adjusted P (Padj) < 0.05; Supplementary Data 1), were enriched for pathways of fatty acid and lipid metabolism, fitting with adipocytic characteristics (Supplementary Table 2).
a, Clinical characteristics of the 15 samples used to build the atlases. Samples were collected from female and male donors with varying age and BMIs, as described in the pie charts. Characteristics were measured directly or extracted from donors’ medical records. b, The expression of the adipocyte marker adiponectin (ADIPOQ) in Uniform Manifold Approximation and Projection (UMAP) representations of hSAT and hVAT snRNA-seq atlases containing 37,879 and 83,731 nuclei, respectively. In both depots, adipocytes constitute a well-defined cluster. c, The expression of cell-specific markers in different clusters of each depot. Adipocytes were distinguished by multiple markers (marked in orange). d, The fraction of adipocytes per depot, stratified by sex, age and BMI in the snRNA-seq in-house data (hSAT, hVAT) and on estimating adipocyte proportions using deconvolution (Decon.) analysis of 73 paired bulk RNA-seq profiles (‘Decon’; hSAT:hVAT). Adipocytes were significantly more prevalent in hSAT versus hVAT in both datasets (two-sided Mann–Whitney U-test P = 0.0013 and P = 1.205 × 10−13, respectively), and tended to be more prevalent in females versus males according to the snRNA-seq data (two-sided Mann–Whitney U-test P = 0.044 in hVAT only). Other differences were not statistically significant. The boxplot central band indicates the median, the box limits the 25th to 75th percentiles and the whiskers 1.5× the interquartile range.
Overall, we identified 12,205 (32.2%) hSAT and 15,460 (18.5% of total) hVAT adipocytes. Adipocytes were more abundant in women than in men in both fat depots and more frequent in hSAT than hVAT, potentially owing to the lower cellular diversity of hSAT (10 cell types, versus 13 in hVAT; Fig. 1d and Supplementary Table 1). These results were corroborated on analyzing our paired hSAT–hVAT samples alone (Supplementary Fig. 1). The larger frequency of adipocytes in hSAT versus hVAT was supported when applying sNucConv—a deconvolution tool tailored for snRNA-seq of adipose tissue16—onto paired hSAT/hVAT bulk RNA-seq data of 73 patients with extreme obesity (Fig. 1d).
To further compare adipocyte populations between depots we applied pseudobulk analysis, followed by differential gene expression between depots (Methods). The hVAT adipocytes were, as expected, enriched for pathways such as insulin resistance (Padj = 0.007), inflammatory mediator regulation of the transient receptor potential channel (Padj = 0.005), adipocytokine signaling pathway (Padj = 0.016) and circadian entrainment (Padj = 0.031; Supplementary Data 2).
We next investigated the diversity of the adipocyte population per depot by subclustering adipocytes based on their snRNA-seq profiles. This analysis revealed seven and eight clusters of hSAT and hVAT adipocytes, which we denoted SA1–7 and VA1–8, respectively (Fig. 2a). Each adipocyte cluster contained nuclei from all samples (Supplementary Table 3 and Supplementary Fig. 2a) and similarly expressed markers of mature (terminally differentiated) adipocytes (ADIPOQ, AQP7, LPL and PLIN1; Fig. 2b). In addition, each adipocyte cluster overexpressed at least 20 genes compared with all other adipocyte clusters (Supplementary Figs. 3 and 4 and Supplementary Data 3), although the degree of overexpression varied (Fig. 2c). For example, the 20 topmost overexpressed genes in the largest cluster per depot, SA1 and VA1 (85% and 75% of the adipocyte nuclei in hSAT and hVAT, respectively), were only modestly overexpressed relative to the 20 topmost overexpressed genes of other clusters (for clarity, Fig. 2c presents only a randomly selected subset of SA1 or VA1).
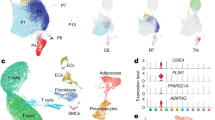
a, UMAP representations of hSAT and hVAT adipocyte atlases containing 12,205 and 15,460 nuclei, respectively. Pie charts show the relative abundance of each adipocyte subset. b, The expression of adipocyte markers in adipocyte clusters of each depot. Distinct clusters expressed markers at similar levels. c, Heatmaps based on the top 20 markers of each adipocyte cluster in hSAT and hVAT. Adipocyte clusters are represented by all their nuclei (columns), except for SA1 and VA1 which are represented by a randomly selected subset of nuclei as a result of their large size. d, Violin plots depicting below-threshold DoubletFinder score (upper) and mitochondrial (MT) content (lower) per adipocyte cluster. Clusters that were validated by immunofluorescence (Fig. 5) were marked with ‘+’. e, UMAP representation of the integrated hSAT and hVAT adipocyte atlases, showing that most subsets overlap between the two depots. f, The proportions of SA1–7 and VA1–8 in each iASV1–7 cluster. The total number of nuclei per integrated cluster appears below the cluster name. g, The number of nuclei from SA1–7 and VA1–8 (columns) that contribute to each integrated cluster (rows). Per column, the entry corresponding to the maximal value (that is, most nuclei) is highlighted.
All adipocyte nuclei passed stringent quality control assessment (detailed in Methods). Yet, when assessing each adipocyte cluster’s subthreshold doublet score, SA2–7 and VA2–5 had higher score distribution compared with SA1 or VA1, and VA6 had a higher mitochondrial gene content (Fig. 2d). To further support the validity of our adipocyte subclusters, we reanalyzed the raw snRNA-seq data using two alternative computational pipelines with enhanced doublet or ambient RNA removal (Supplementary Note). Subclustering of the identified adipocytes showed that adipocyte clusters were retained in both pipelines, mostly as distinct clusters (Supplementary Figs. 5 and 6).
To compare hSAT and hVAT adipocyte landscapes, we combined our depot-specific datasets, reclustered them into depot-integrated adipocyte clusters and assessed the composition of each integrated cluster (denoted ‘integrated adipocytes subcutaneous and visceral‘ (iASV); Fig. 2e and Supplementary Table 4a). Notably, SA1–5 and VA1–5 contributed to iASV1–5, respectively, demonstrating the similarity in adipocyte subpopulations of the two depots. For example, >98% of SA1 nuclei and >99% of VA1 nuclei clustered together into iASV1 (Fig. 2f,g). We also observed depot differences, because iASV6 was contributed predominantly by VA6 (68% of iASV6 cells).
Characterizing adipocyte subpopulations
To infer the putative functions of the different adipocyte clusters, we subjected SA1–7 and VA1–8 separately to functional enrichment analysis, based on their differential expression profiles relative to other adipocyte clusters in the same depot (Methods, Supplementary Data 4 and Fig. 3). In both depots, SA1 and VA1 clusters were enriched for fatty acid and lipid metabolic processes and were therefore denoted as ‘classical adipocytes’ (Fig. 3b). Their classical adipocytic functions, relatively common to all adipocyte clusters, explain their modest overexpression pattern observed in Fig. 2c. Most of the remaining clusters were enriched for nonclassical adipocyte functions: ‘angiogenic adipocytes’ (SA2,VA2) were enriched for angiogenesis, blood vessel and vascular development processes (Fig. 3c). ‘Immune-related adipocytes’ (SA3,VA3) were enriched for immune-regulation processes, such as ‘positive regulation of phagocytosis’ and ‘macrophage activation’ (Fig. 3d). Additional immune-related adipocytes (SA4,VA4) were also enriched for lymphocytic processes, including ‘T cell activation’ and ‘adaptive immune response’ and were thus named ‘immune-related (adaptive) adipocytes’ (Fig. 3e). ‘Extracellular matrix (ECM) adipocytes’ (SA5,VA5), were enriched for ECM processes, including ‘ECM structural constituent’ and ‘ECM organization’ (Fig. 3f). The depot-specific cluster VA6 was enriched in mitochondrial and ribosomal genes and processes (for example, ‘mitochondrion organization’, ‘mitochondrial respiratory chain complex I assembly’, ‘ribosomal subunits’, ‘protein targeting to endoplasmic reticulum’, Supplementary Data 4) and were thereby termed ‘mito-ribosomal adipocytes’ (Fig. 3g). Clusters SA6–7 in hSAT and VA7–8 in hVAT, which contained small numbers of nuclei, were not significantly enriched for specific processes and remained unannotated. Given the SA1/VA1 distinction from the remaining adipocyte clusters, we collectively named the latter ‘nonclassical adipocytes’.
a, UMAP representations of the in-house hSAT and hVAT adipocyte atlases and the combined hSAT and hVAT adipocyte atlas of Emont et al.9. Each UMAP is colored by the adipocyte subtypes defined herein (for hSAT and hVAT) or by Emont et al. (right). b–g, UMAP representations of hSAT, hVAT and Emont et al. adipocyte atlases: lipid metabolism (b); angiogenesis (c); innate immune (d); adaptive immune (e); ECM (f); and mito-ribosomal (g). Each cluster is colored by its mean normalized enrichment scores (NES) for GO annotations related to the process specified in the panel (see color legend at bottom right). Adipocyte subtypes were enriched for distinct processes, suggesting that they have different functionalities.
We next searched for potentially active regulons (transcription factors and their target genes) per adipocyte subtype using SCENIC17 (Methods, Supplementary Table 5 and Fig. 4a). Certain regulons were active in all adipocyte subtypes, including key adipogenesis transcription factors (ATF2, CEBPB), consistent with the similar expression of fully differentiated adipocyte markers in all adipocyte subtypes from both depots (Fig. 2b). Depot differences were also noted. For example, CEBPA regulon was identified in VA1–8, but not in hSAT adipocyte subtypes. Other regulons were unique to the different nonclassical adipocyte subtypes in both depots, some supporting the putative adipocyte subtype functions. For example, PRRX1, which characterized the ECM subtype VA5, is known to regulate transforming growth factor-β (TGFB)-mediated fibroblast-to-myofibroblast differentiation, fibroblastic phenotype of adipocytes18 and expression of COL6A3 (ref. 19). Likewise, MEF2C and the nuclear factor-κB-subunit RELB characterized the immune subtypes in both depots (SA3–4, VA3–4). Some regulons, occasionally of the same family of transcription factors, were depot specific: of the cytokine-response STAT family, STAT4 characterized SA4 whereas STAT6 characterized VA4. Jointly, the regulon analysis provided a depot similar and differential regulatory network view, supporting the putative functionality of nonclassical adipocytes.
a, Potentially active regulons (rows) within adipocyte subtypes (columns). The regulons were named by their transcription factor; circle size and color intensity indicate the percentage of nuclei with a potentially active regulon. The gray boxes mark all adipocyte subtypes (top box) or specific subtypes. b,c, UMAP representations of the integration of our in-house adipocyte atlases (green, hSAT; pink, hVAT) and the atlases of Emont et al.9 (gray): i-SAT (b) and i-VAT (c). The proportions of SA1–7 and VA1–8 in each integrated SAT (i-SA1–7) or VAT (i-VA1–5) cluster are depicted in the bar plots. The total number of nuclei per integrated cluster appears below the cluster name. d,e. The number of nuclei (columns) from SA1–7 (d) or VA1–8 (e) and Emont et al. that contribute to each integrated cluster (rows). Per column, the entry corresponding to the maximal value (that is, most nuclei) is highlighted. f, The similar distribution of nuclei among nonclassical adipocyte clusters for the in-house dataset (SA2–7, left) and the Emont et al. atlas (i-SA2–7). g, Same as f, for hVAT (VA2–8, i-VA2–5).
Last, we tested whether nonclassical adipocytes were present in other atlases and datasets of human adipose tissues. We first focused on the atlas of Emont et al.9, which contained 14,396 hSAT and 11,475 hVAT adipocytes. To assess the similarity among adipocytes of each atlas, we integrated them separately for hSAT and hVAT and assessed the composition of each integrated cluster (i-SA in SAT, i-VA in VAT; Fig. 4b–g, Supplementary Table 4b,c and Supplementary Fig. 7). In hSAT, we identified seven integrated clusters (Fig. 4b). Although Emont et al. contributed more hSAT adipocytes than our in-house data, i-SA1–7 reassuringly matched our in-house clusters SA1–7 (Fig. 4d). Most (92.7%) of the Emont et al. adipocytes appeared in the integrated classical adipocyte i-SA1 cluster. The remaining adipocytes (7.3%) segregated into each of the remaining integrated clusters, with each cluster containing ten to hundreds of nuclei distributed similarly to our in-house data (Fig. 4d,f and Supplementary Table 4b). In hVAT, we identified five integrated clusters (i-VA1–5) that matched our in-house clusters VA1–5 (Fig. 4c; the small VA6–8 were integrated into i-VA1). As in hSAT, most (86.5%) hVAT adipocytes of Emont et al. appeared in the classical i-VA1 cluster, whereas the remaining 13.5% split into each of the other nonclassical adipocyte clusters (Fig. 4e,g and Supplementary Table 4c). A similar view was obtained with riverplots of the two atlases (Supplementary Fig. 8). As an additional test for the existence of nonclassical adipocytes in Emont et al.’s atlas, we trained subtype-specific classifiers on our in-house adipocyte subtypes and applied them to Emont et al.’s adipocytes (Methods and Supplementary Fig. 9a). This analysis also revealed considerable fractions of nonclassical adipocytes per depot (Supplementary Fig. 9b and Supplementary Table 6). Last, functional enrichment analysis of Emont et al.9 adipocyte clusters hAd1–7 showed enrichment for the nonclassical adipocyte functions (Supplementary Data 4 and Fig. 3c–g).
The presence of nonclassical adipocytes was also observed in the hSAT spatial transcriptomics dataset of Backdahl et al.11 and on applying sNucConv deconvolution16 to the 73 paired hSAT/hVAT bulk RNA-seq profiles, at expected proportions (Supplementary Note and Supplementary Figs. 9c and 10). Thus, multiple computational analyses and datasets support the presence of classical and nonclassical adipocytes in hSAT and hVAT.
Microscopic detection of nonclassical adipocytes
Next, nonclassical adipocytes were validated directly at the protein level in histological sections of human adipose tissues. We chose PTPRB, PDE4D, SKAP1 and ANK2 as markers to identify SA2–5/VA2–5, respectively (Methods and Fig. 5a–d). As each marker was also expressed in nonadipocyte cell types (Extended Data Fig. 1), we also stained for the adipocyte protein perilipin 1 (PLIN1). Immunofluorescence analyses were performed on paraffin sections of hSAT and hVAT using marker-specific commercial antibodies (Supplementary Table 7). To ensure specificity of the fluorescent signal, each antibody was first tested on a positive control tissue. Fluorescence emission spectra from these stained sections were comparable to positive signals from stained adipose tissue, as were nonprimary antibody negative controls (Extended Data Fig. 2). Nonclassical adipocytes identified as being PLIN1 plus subtype-specific, double-positive markers were identified, confirming the presence of SA2–5/VA2–5 within hSAT and hVAT (Fig. 5e–h and Extended Data Fig. 3). Nonclassical adipocytes were dispersed in the tissue, relatively rare among total PLIN1+ adipocytes and without clearly discernable localization within adipose lobules, consistent with the dataset analysis of Backdahl et al.11 (Supplementary Fig. 10).
a–d, UMAP representations of hSAT (left) and hVAT (right) adipocyte subpopulations depicting the expression of specific markers used to identify each subpopulation of nonclassical adipocytes. The markers used were PTPRB (a), PDE4D (b), SKAP1 (c) and ANK2 (d), corresponding to the identification of SA2/VA2–SA5/VA5, respectively. The red arrow indicates the specific nonclassical adipocyte subpopulation represented by the marker. e–h, Representative images from both hSAT and hVAT of adipocytes displaying positive signal for the corresponding marker, visualized using immunofluorescence staining for PLIN1 (green), DAPI (blue), PTPRB (e; red), PDE4D (f; red), SKAP1 (g; yellow) and ANK2 (h; magenta). Scale bar, 20 μm. In f, the arrow indicates the PDE4D-positive adipocyte and the arrowhead points to a crown-like structure (CLS). Representative images of adipocytes from six to eight different patients (from at least two independent staining experiments) are shown. i–k, Validating VA6 in hVAT. i, Violin plots of nuc-MT gene expression in the hVAT adipocyte clusters VA1–8. VA6 had significantly higher expression of nuc-MT genes relative to other hVAT adipocytes (two-sided Mann–Whitney U-test P < 2.2 × 10−16). j, Expression of MT-CO2 in hVAT adipocyte-clustering UMAP (arrow pointing to the relatively highly expressing VA6). k, Immunofluorescence staining for CD36 (green), MT-CO2 (red) and DAPI (blue). *Adipocytes identified with CD36 rather than PLIN1 to enable co-staining with MT-CO2. Scale bar, 20 μm. Representative images of adipocytes from five different patients (two independent staining experiments) are shown.
The hVAT-specific mito-ribosomal adipocyte subtype (VA6) was dubious owing to expression of mitochondrial genes. Yet, consistent with ref. 20, the hVAT adipocyte exhibited higher expression of nuclear-encoded mitochondrial (nuc-MT) genes compared with hSAT, with VA6 showing the highest expression (Fig. 5i and Supplementary Fig. 11). Immunostaining hVAT for the VA6 mitochondrial marker protein, MT-CO2 (Fig. 5j), revealed rare MT-CO2-positive adipocytes (positively stained for CD36 instead of perilipin, for technical reasons; Fig. 5k and Extended Data Figs. 2e and 3d). These analyses suggest that VA6 could represent a rare, bona fide hVAT nonclassical adipocyte subtype characterized by high mitochondrial content. Collectively, complementary approaches, different datasets and assessment of protein expression of RNA-based adipocyte subtype markers support the identification of classical and nonclassical adipocytes in the two human fat depots.
To determine whether classical and nonclassical adipocytes correlate with clinical characteristics, we used their sNucConv estimated proportions per sample in the 73 paired hSAT/hVAT bulk RNA-seq profiles of patients with severe obesity (Extended Data Fig. 4). Whereas classical adipocytes’ proportions in hSAT exhibited no correlation with body fat, their higher proportions correlated with lower waist:hip ratio and, consistently, with lower insulin resistance (assessed by the homeostatic model of insulin resistance). Conversely, higher hSAT nonclassical adipocyte proportions correlated with greater insulin resistance, an effect that trended similarly in hVAT although without reaching statistical significance. Higher hVAT classical adipocytes proportions tended to correlate with lower fasting triglyceride levels and predicted a smaller decline in triglycerides 1 year post-bariatric surgery. This is consistent with studies demonstrating greater metabolic benefits of bariatric surgery in patients with more severe, preoperative, obesity-related metabolic dysfunction. Predicting bariatric surgery outcome by preoperative hVAT classical adipocyte proportions highlights the potential for using estimated or directly assessed adipocyte subtype abundance in the clinic.
Differentiation trajectories toward adipocyte subtypes
Adipose tissues harbor a large and diverse population of mesenchymal stem cells with differentiation potentials to a range of terminal differentiation states. The subpopulation of cells most committed to become mature adipocytes are termed preadipocytes and the entire cell population is characterized by expressing PDGFRA, and termed adipose stem and progenitor cells (ASPCs9 or fibroadipogenic progenitors10). To investigate potential differentiation paths per depot from ASPCs toward classical and nonclassical adipocyte subtypes, we applied trajectory analysis tools21,22.
In hSAT, ASPCs were identified in a PDGFRA-positive cluster, comprising 26% of the nuclei (Fig. 6a), and could be divided into five subclusters, ASPC1–5, marked by known ASPC subtype markers (Fig. 6b-c). ASPC1 (32% of the ASPCs) expressed markers of highly proliferative multipotent progenitor cells9, including DPP-4 (ref. 23) and CD55 (ref. 10) and was F3 negative23 (referred to as adipose stem cells in ref. 8). ASPC2 (55%) expressed peroxisome proliferator-activated receptor-γ (PPARG) most highly and was CD9low, suggesting its commitment to adipocyte fate24. ASPC3 (9%) expressed markers of profibrotic progenitor cells (CD9high), previously observed and designated myofibrogenic progenitors in humans and mice23,24. ASPC4 expressed PDE4D most highly.
a, The expression of the ASPC marker PDGFRA in UMAP representations of hSAT snRNA-seq atlases. ASPCs constitute a well-defined cluster. b, The proportion of ASPC subclusters within the hSAT ASPCs. c, The expression of ASPC markers and of stem, fibrosis and adipogenic markers in hSAT ASPC subclusters. d, ASPCs, classical and nonclassical adipocytes in hSAT colored by cell group (left) with inset showing ASPCs colored by subcluster. Right, nuclei colored by their differentiation pseudotime from ASPCs (ASPC1). Nonclassical adipocytes were an intermediate state between ASPCs and classical adipocytes. Visualization was performed using ForceAtlas2 (FA) representation. e,f, Same as a (e) and b (f) for hVAT. g, The expression of ASPC markers and stem, fibrosis, adipogenic and mesothelial markers in hVAT ASPC subclusters. h, Same as d for hVAT. Nonclassical adipocytes appear as an intermediate state between ASPCs and classical adipocytes. ASPC4 shows a pseudotime comparable to nonclassical adipocytes, suggesting that it might lead to a different end-state.
As expected, a clear pseudotime path connected hSAT ASPCs to adipocytes (Fig. 6d): the proliferative ASPC1, which was the starting state for this analysis, and the profibrotic ASPC3, which was the most distant from adipocytes, whereas ASPC2 and ASPC4 were the closest. Importantly, classical adipocytes were the probable terminal differentiation state via nonclassical adipocytes. This suggests that the classical adipocyte differentiation state stems from loss, rather than gain, of nonclassical adipocytes’ functions.
In hVAT, PDGFRA-positive hVAT-ASPCs (Fig. 6e) were divided into four subclusters (ASPC1–4), sharing markers and integrated nicely with hSAT-ASPC1–4 (Fig. 6f,g and Supplementary Fig. 12b). Trajectory analysis showed that, as in hSAT, the proliferative (ASPC1) and the profibrotic (ASPC3) clusters were the most distant from adipocytes, whereas the committed (ASPC2) clusters were closer (Fig. 6h). Furthermore, as in hSAT, hVAT-ASPCs linked to classical adipocytes via nonclassical adipocyte subtypes (Fig. 6h).
In both depots, trajectory analysis using an alternative tool, alternative starting points and a stringently filtered dataset consistently showed that classical, rather than nonclassical, adipocytes, are an ASPC differentiation end-state (Supplementary Note and Supplementary Figs. 12–14). Furthermore, supporting the ASPC → nonclassical adipocytes → classical adipocytes differentiation path were the result of ImageStream flow cytometry analyses, recently shown to enable capturing both stromal cells and adipocytes25. PTPRC protein (CD45, common leukocyte antigen), the mRNA of which was preferentially expressed in SA3–4/VA3–4 immune-related, nonclassical adipocytes (not shown) was observed by both immunohistochemistry and immunofluorescence (Extended Data Fig. 5a–d). Moreover, immunostaining of CD45 followed by flow cytometry readily identified stromal vascular fraction cells, potentially macrophages and CD45− adipocytes (based on round morphology, diffuse Bodipy neutral lipid staining, CD36+, nuclear Draq5− (ref. 26) and with typically polarized light scattering; Extended Data Fig. 5e). Using the same criteria, ~7% of cells had membrane-appearing CD45 staining in adipocytes, which typically were of small size. The presence of particularly small-sized PTPRC/CD45+ adipocytes is consistent with the notion that nonclassical adipocytes are an intermediate between adipocyte precursor cells and full-sized, lipid-laden, terminally differentiated, CD45−, classical adipocytes.
Depot-specific adipocyte communication routes
Adipose tissues are key endocrine organs relying on adipocyte paracrine communication. We first examined the adipocyte-generated hormones adiponectin and leptin. Both were expressed similarly across hSAT and hVAT adipocyte subtypes (except that leptin was largely missing from VA7; Extended Data Fig. 6a). Next, we examined their receptors: adiponectin signals through ADIPOR1, ADIPOR2 and CDH13 (T-cadherin), whereas leptin uses LEPR. In both depots, ADIPOR1 was largely absent from all adipocyte subtypes, ADIPOR2 was ubiquitous, CDH13 was expressed uniquely by angiogenic adipocytes (SA2/VA2) and LEPR was expressed in most subtypes (Extended Data Fig. 6a). In nonadipocytes, ADIPOR2 expression was detectable only in a few hVAT cell types, although, like mast cells and endothelial cells, they were present in both depots (Extended Data Fig. 6b). It is interesting that CDH13 expression characterized endothelial cells in both depots (although lymphatic endothelial cells expressed CDH13 only in hSAT), suggesting that adiponectin may similarly affect angiogenic adipocytes and endothelial cells.
We explored additional, adipocyte subtype, communication routes. CellphoneDB27 analysis showed that communication among adipocytes was more active in hSAT than hVAT (212 versus 110 median interactions, respectively; Supplementary Data 5 and Fig. 7a). Next, we estimated communication probabilities among adipocyte subtypes for specific interadipocyte ligand–receptor pairs using CellChat28, which also suggests ‘key’ incoming and outgoing signaling pathways (Fig. 7b–i). Adiponectin signaling was a key pathway in both depots (Fig. 7b). Classical adipocytes (SA1/VA1) were the main senders of adiponectin in both depots, possibly reflecting their dominant proportions among the various adipocyte subtypes (Fig. 7b). Leptin signaling was a key pathway only in hVAT (Fig. 7c), consistent with low LEPR expression in SA1 (Extended Data Fig. 6a and Supplementary Data 5). This suggests that, although hSAT is the major source of leptin production29,30, its classical adipocytes might be less leptin sensitive.
a, The total number of interactions between cell types in hSAT and hVAT. The number of interactions among adipocyte subtypes is higher in hSAT. The number of interactions between adipocytes and the macrophage or myeloid compartment or mast cells is higher in hVAT compared with hSAT. The heatmap was calculated using CellPhoneDB. b–i, The senders and receivers of specific signaling pathways among adipocytes in hSAT and hVAT. Analysis was performed using CellChat. b, classical adipocytes as the main senders of adiponectin in both depots. c, Leptin pathway missing from hSAT. In hVAT, classical adipocytes VA1 and VA7,VA8 are the main senders and VA2 and VA5 the main receivers. d,e, In both depots, angiogenic adipocytes SA2,VA2 are the main receivers of the angiogenesis-related ANGPT (d) and VEGF (e) signaling pathways. f, In both depots, immune-related adipocytes SA4,VA4 and SA3,VA3 are the main senders and receivers, respectively, of the immune CD45 signaling pathway. g, In hSAT alone, the anti-inflammatory signaling pathway Annexin is received mainly by SA3. h, In hVAT alone, the proinflammatory signaling pathway IL-16 received mainly by VA3. i, In both depots, the ECM adipocytes SA5,VA5 are the main senders of the ECM-related laminin signaling pathways.
Other signaling pathways revealed several subtype-specific patterns. In both depots, angiogenic adipocytes (SA2/VA2) were the most likely adipocyte subtype to be targeted by the angiogenesis-related angiopoietin (ANGPT) and vascular endothelial growth factor (VEGF) signaling pathways (Fig. 7d,e). Immune-related adipocyte subtypes (SA3–4/VA3–4) similarly exhibited the immune pathways CD45 (Fig. 7f) and colony stimulating factor (CSF; Supplementary Data 5) as key signaling pathways. However, although the hSAT SA3 adipocytes were targeted by the anti-inflammatory annexin (Fig. 7g) and galectin pathways (Supplementary Data 5), their respective hVAT VA3 equivalents were targeted by the proinflammatory interleukin (IL)-16 key pathway (Fig. 7h). This is consistent with the notion of hVAT being more inflammatory than hSAT, suggesting that these adipocyte subtypes participate in mediating this depot difference. ECM adipocytes (SA5/VA5) were the main senders of the laminin pathway, which plays major roles in adipose tissue ECM deposition31, in both depots (Fig. 7i).
We further explored whether the hSAT–hVAT difference in inflammatory communication extends beyond adipocytes by also considering the adipose macrophage or myeloid compartment (including bona fide macrophages, dendritic cells, monocytes and pre-myeloid cells). In hSAT, adipocyte subtype interactions with this cell compartment were clearly less pronounced relative to intra-adipocyte interactions (two-sided Mann–Whitney U-test P < 10−9; Fig. 7a). In hVAT, in contrast, adipocyte interaction with subclasses of this compartment (pre-myeloid 1 and 2) seemed to be more intense (two-sided Mann–Whitney U-test P = 0.002; Fig. 7a). Similarly, in hVAT, mast cells interacted more with adipocytes and myeloid cells than in hSAT (Fig. 7a). To further explore this seemingly enhanced proinflammatory pattern in hVAT compared with hSAT, we examined the IL-16 and annexin pathways on including the macrophage or myeloid compartment. The proinflammatory IL-16 signaling involved many cell types of the macrophage or myeloid compartment in both depots and with similar intensity, except for the unique role of VA3 as the sole adipocyte subtype receiver (Supplementary Data 5I,K). The anti-inflammatory annexin signaling was much more pronounced in hSAT versus hVAT and involved many more cell types as senders or receivers (Supplementary Fig. 15 and Supplementary Data 5). We also observed low interaction intensity between adipocytes and mast cells in hSAT compared with hVAT (Fig. 7a). Mast cells were targeted by adiponectin in hVAT but not in hSAT (Extended Data Fig. 6b) and, as source, were highly involved in ECM, fibrosis and IL-16 pathways in hVAT (Extended Data Fig. 7). Thus, depot-differential communication patterns between adipocyte subtypes and mast cells may also contribute to the differential inflammatory and fibrotic ‘tone’ between the two fat depots.
Discussion
snRNA-seq provides unprecedented resolution of the cellular landscape of human adipose tissues9,10,11,12,13,15. It is interesting that, although some fat depot differences were observed9,10,12,15, snRNA-seq profiles of hSAT and hVAT were largely similar, despite known functional and whole-tissue transcriptomic differences. In the present study, we hypothesized that in-depth, depot-specific analyses of adipocyte atlases obtained using snRNA-seq could elucidate some of the differences between human fat depots. Hence, we identified adipocyte subtypes (characterized by proposed functionality), differentiation trajectories from ASPCs, and intercellular communication patterns.
Both hSAT and hVAT contain mostly classical adipocytes and several nonclassical subtypes with unique putative functions (Fig. 3). As previously noted9,10,15, the depots’ adipocyte subtype composition is quite similar (Fig. 2a). Process enrichment analyses provided clearer functional annotation of these subpopulations than single-gene markers (Fig. 3b–g). In both depots, classical adipocytes, which mildly preferentially expressed lipid metabolism and handling genes, constituted the largest adipocyte subtypes (Fig. 2a). Previously, three hSAT adipocyte subtypes were identified by different expression levels of LEP, PLIN1 and SAA, with one subtype more transcriptionally responsive to in vivo insulin stimulation11. Although our data cannot assess insulin sensitivity, marker genes used herein suggest that their parallel adipocyte subtypes probably fall within the large, classical adipocyte cluster in our dataset.
The next-sized, nonclassical adipocyte subtypes (with depot-different abundances) were ECM-related adipocytes, angiogenic adipocytes and immune-related adipocytes. The ECM subtype, SA5/VA5, enriched in ECM genes, also expressed PDGFRB, possibly relating it to fibroinflammatory progenitors previously reported in mice32. Yet, importantly, all adipocyte subtypes in our study similarly expressed LEP, ADIPOQ, PLIN1 and other adipocyte-unique genes, attesting to their mature adipocytic character. Depot-specific adipocyte subtypes were identified in ref. 9 and, consistently, a small adipocyte subtype, VA6, was observed in our dataset in hVAT alone. VA6 was characterized by relatively high expression of mitochondrial and ribosomal genes or functions, possibly consistent with a high protein translation activity and/or a high mitochondrial content, which characterizes hVAT20. Finally, mature adipocytes and nuclei that express both adipocyte and macrophage markers (MHCII and CD45) were also reported in mice25,33 and humans25, consistent with our SA3–4 and VA3–4. It is interesting that active regulon analysis by SCENIC revealed that similar putative functions of nonclassical adipocytes may be achieved in hSAT and hVAT via partially distinct regulons, occasionally involving different members of same-family transcription factors (Fig. 4a).
Nonclassical adipocyte subtypes expressed genes that are more identified with other cell types (Extended Data Fig. 1). This prompted us to utilize several complementary computational and experimental approaches to rule out doublets or ambient RNA. First, we repeated the computational analyses using alternative pipelines that stringently filtered out putative doublets and ambient RNA and obtained similar results (Supplementary Note). Second, VA6, despite high mitochondrial gene content (frequently considered as low quality, Fig. 2d), also showed the highest nuclear-encoded mitochondrial gene expression (Fig. 5i), suggesting that it is a true hVAT-specific adipocyte subtype with high mitochondrial content. Third, we found multiple evidence for nonclassical adipocytes in additional human adipose tissue datasets, processed independently of our computational pipeline. Finally, and most importantly, we utilized immunofluorescence analyses to identify nonclassical adipocytes that express putative marker proteins predicted by the snRNA-seq data, in histological sections (Fig. 5 and Extended Data Figs. 1–3 and 5). Notably, each antibody identified within adipose tissue also stained nonadipocyte cells known to express their respective protein, providing an internal positive control. We verified the identity of these markers’ expression within adipocytes by co-staining for PLIN1, a common adipocyte marker (or CD36 for VA6). All nonclassical adipocytes were not highly abundant in the tissue and represented a low percentage of total adipocytes, largely consistent with the snRNA-seq analysis. Jointly, although the possibility of doublets or low-quality nuclei cannot be fully excluded, our complementary approaches suggest that nonclassical adipocytes are bona fide adipocytes of human adipose tissues, identifiable at both the nuclear mRNA and the whole-cell protein level.
Trajectory analysis from ASPCs to adipocytes in both depots suggested that classical adipocytes are the terminal differentiation state, via nonclassical adipocytes (Fig. 6 and Supplementary Figs. 12–14). Thus, unique functions of nonclassical adipocytes may be ‘lost’ during biogenesis of classical adipocytes. Both hSAT and hVAT adipocytes probably originate from PPARG-expressing ASPCs. Experimental proof for these differentiation trajectories, such as using lineage tracing approaches, are unattainable in human adipose tissue samples. Similarly, in vitro differentiation of human adipose-derived stromal vascular cells into adipocytes requires combinations of extremely high, supraphysiological concentrations of hormones and signaling pathway activators. These generate a highly artificial environment that fails to reproduce a versatile human adipocyte population, even well-characterized adipocyte subtypes, such as beige adipocytes. Future mechanistic studies will therefore have to rely on model systems more amenable to genetic manipulation and lineage tracing.
As for intercellular communication patterns, hSAT adipocyte subtypes were predicted to highly interact, more than with adipose tissue immune (mostly myeloid) cells, whereas this is the opposite in hVAT (Fig. 7a). Moreover, adipocyte subpopulations common to the two depots, such as immune-regulating adipocytes (SA3/VA3), participate in intercellular communication through depot-different dominant interaction pathways (Fig. 7g,h). These provide new ways to understand the more proinflammatory nature of hVAT over hSAT.
Our study has several noteworthy weaknesses and strengths. As in other single-cell studies, there is the limited number of samples assessed (high-cost technology) and, indeed, the meta-analysis of data from different centers has recently been published10. We employed several approaches to address this shortcoming. First, donors present diversity of sex, age and BMI (Fig. 1a), while carefully selected to exclude severely morbid conditions. We used multiple approaches to compare our adipocyte dataset with that of Emont et al.9, to verify that adipocyte subtypes in our data are also identifiable in an independent dataset (Figs. 3 and 4b–g). We used deconvolution to estimate classical and nonclassical adipocytes' proportions in a larger cohort (Supplementary Fig. 9) and immunofluorescence to identify nonclassical adipocytes in tissue sections using protein markers. Another important aspect is our separate analysis of each depot, particularly of the adipocyte population. Although combined hSAT/hVAT analysis could enhance signals that might otherwise remain undetected, it could also mask depot differences. Our depot-independent analysis enabled detecting commonalities and some more subtle differences in an unbiased fashion. These were then verified using additional pipelines for snRNA-seq processing, machine learning classification and deconvolution and additional datasets, including other snRNA-seq and bulk RNA-seq profiles of human adipose tissue.
In conclusion, separate analyses of hSAT and hVAT refines understanding of commonalities and differences in adipocyte cell composition, differentiation pathways and intercellular communication between human fat depots. This characterization offers a valuable resource for future mechanistic and clinically oriented studies.
Methods
Collection of human samples
Adipose tissue biopsies weighing at least 1 g were obtained from adult donors without severe morbid states or obesity complications who had signed, in advance, a written informed consent before undergoing elective abdominal surgeries. For 14 samples, ethical approval of the study procedures was obtained by the Helsinki Ethics Committee of Soroka University Medical Center (approval no. 15-0348). Donors were not compensated for agreeing to participate in the study. For one sample, collection was approved in the context of the Leipzig Obesity BioBank (LOBB; n > 8,000 donors; BMI range 13–29 kg m−2; age range: 18–96 years) by the Ethics Committee of the Medical Faculty of the University of Leipzig (approval nos. 159-12-21052012 and 017-12-ek) and performed in accordance with the Declaration of Helsinki. Five donors donated hSAT and hVAT samples (paired). Clinical characteristics of donors were measured directly or extracted from medical records (Fig. 1a and Supplementary Table 1). Biopsies were immediately delivered from the operating room to the lab, where they were snap-frozen in liquid nitrogen and stored at −80 °C, as detailed previously34,35.
SnRNA-seq profiling
Human adipose tissue samples were analyzed by isolating nuclei, barcoding their transcriptome using Chromium 10x technology, generating complementary DNA and then expression libraries, followed by sequencing of the expression libraries. Briefly, nuclei were isolated from 500 mg of frozen hSAT or hVAT samples with all steps carried out on ice. Then, 1 ml of ice-cold adipose tissue nuclei lysis buffer (AST: 5 mM PIPES, 80 mM KCl, 10 mM NaCl, 3 mM MgCl2 and 0.1% IGEPAL CA-630) supplemented with 0.2 U µl−1 of RNase Protector (Roche) was added to each sample, followed by mincing the frozen tissue using small surgical scissors. Then, small pieces of minced tissue in 1 ml of AST lysis buffer were transferred into 7 ml of WHEATON Dounce Tissue Grinder (DWK Life Sciences) and an additional 2 ml of AST was added. Tissues were dissociated to retrieve nuclei by 12 strokes with a loose pestle, followed by 10 strokes with a tight pestle. Samples were then incubated for 7 min on ice to ensure maximal nuclei retrieval, filtered through a 70-µm SmartStrainer (Miltenyi Biotech) and centrifuged (4 °C, 5 min, 500g). Supernatants were discarded and the nuclei pellet was resuspended in 0.5 ml of ice-cold phosphate-buffered saline (PBS) + 0.5% bovine serum albumin (BSA), supplemented with 0.2 U µl−1 of RNase Protector and filtered through a 40-µm SmartStrainer, followed by additional centrifugation. Next, 2 ml of the supernatant was discarded and the nuclei pellet was resuspended in the remaining 0.5 ml of ice-cold PBS + 0.5% BSA. Nuclei were counted using LUNA-FL Dual Fluorescence Cell Counter (Logos Biosystems) and 13,000–16,500 nuclei per sample were immediately loaded on to 10x Chromium Controller (10x Genomics) according to the manufacturer’s protocol (Chromium Next GEM Chip C. Chromium Next GEM Single Cell 3ʹ-Kits v.3.1). The cDNA and gene expression libraries size fragments were assessed using 4150 TapeStation System (Agilent) with high-sensitivity D5000 ScreenTape and high-sensitivity D1000 ScreenTape, respectively, while their concentrations were quantified using Qubit 4 Fluorometer using Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific). Gene expression libraries were sequenced by NextSeq 550 using High Output Kit v.2.5 (150 cycles) kit and NovaSeq 6000 using S1 100 cycles flow cell kit at a sequencing depth of at least 50,000 reads per nucleus.
SnRNA-seq data pre-processing and quality control
Per sample, reads were transformed into a raw counts matrix with 10x Genomics CellRanger software1 v.6.0.1, which was used as input for CellBender36 v.0.2.0 to remove ambient RNA. Saturation plots appear in Supplementary Fig. 16. For downstream analysis, only cell barcodes that were determined to be true cells by both CellRanger and CellBender algorithms were used. The resulting raw counts matrix of each sample was analyzed with the Seurat v.3.0R package37,38. Our quality control procedure included removal of broken nuclei, that is, nuclei with <200 genes or nuclei that contained >20% mitochondrial genes. Counts were normalized and scaled using the 3,000 most variable genes. Linear dimensionality reduction via principal component analysis and graph-based clustering via k-nearest-neighbors (k = 20) methods were used to group individual cells into cell subsets. To remove plausible doublets, we removed cell clusters with low mean gene count and low mean reads counts (<1 standard deviation below the sample’s mean) and then applied DoubletFinder R package39 v.2.0.4. All cells that were marked as doublets by DoubletFinder were removed, along with cell subsets that contained >65% doublets. Once each sample passed quality control, we integrated all samples using Harmony40 v.1.2.1 and performed dimensionality reduction and clustering again. Clusters were calculated with 0.8 resolution. To avoid over-clustering, clusters were tested with AssessNodes function from Seurat v.2.0, which we restored to fit Seurat v.3.0 objects. Clusters that were not significantly different were merged (P < 0.05). To avoid sample-specific clusters, clusters derived mostly from a single sample (>80% of cluster cells) were not considered in subsequent analyses (Supplementary Fig. 2).
Identification of cell clusters and cluster annotation
Differentially expressed marker genes were calculated using Seurat v.3.0 FindAllMarkers function (logfc.threshold = -inf and min.pct = 0). Cell subsets were annotated using known markers from the literature (such as ADIPOQ and AQP7 for adipocytes), expert knowledge and CellTypist R package41 v.2. Detailed description of the annotation of nonadipocytes appears in ref. 16.
Adipocyte subtype detection and characterization
Adipocyte clusters were extracted from the atlas of each depot and subclustered in a depot-specific manner, including normalization, determination of variable genes, scaling and integration using Harmony40. Nuc-MT genes were obtained from MitoCarta v.3.0 (ref. 42). To characterize adipocyte subtypes, we used gene-set enrichment analysis (GSEA) with clusterProfiler43 v.4.10.1 and gene ontology (GO) as reference gene sets and the Gorilla web server44. We then associated an adipocyte subtype with a subgroup of processes for which it was significantly enriched (Padj < 0.05; the selected processes appear in Supplementary Data 4). For visualization of the enrichment for a given process subgroup, we calculated per subtype the mean of the GSEA normalized enrichment score (NES) for this subgroup (if no enrichment was detected, the NES value was set to 0). Cells within a subtype were then colored by the calculated mean score of the subtype. To identify potentially active transcription factors, we used SCENIC v.1.1.2 (ref. 17). To assess the sampling coverage of adipocyte clusters, we computed cluster preservation in down-sampled datasets45. Specifically, we implemented a procedure that randomly removes 1,000 adipocyte nuclei, clusters the remaining nuclei and checks whether the original adipocyte clusters were preserved. This procedure was repeatedly applied until the number of nuclei was <1,000 and the entire process was conducted 10× per depot. As shown in Supplementary Fig. 17, saturation was observed when the number of nuclei exceeded ~6,400, suggesting that adipocyte clusters represent most of the population present in each depot under physiological conditions. We applied a similar analysis to cell types of the entire depot, randomly removing ~10% of the initial number of nuclei per run (Supplementary Fig. 17). Last, we tested the robustness of adipocyte subtype detection by applying alternative snRNA-seq pre-processing and quality control pipelines, as described in Supplementary Note and Supplementary Figs. 5 and 6.
Comparison between in-house hSAT and hVAT adipocytes
To test the similarity between the adipocytes of each depot, we created a combined adipocyte dataset that included all adipocytes from hSAT and hVAT. The combined dataset was reanalyzed, including normalization, determination of variable genes and scaling, and integrated using Harmony40 with the grouping variable set to sample name and depot. To test for depot-specific functional differences, we applied enrichment analysis to pseudobulk profiles. Specifically, per sample, we created a pseudobulk expression profile by extracting its adipocyte nuclei and aggregating the counts per gene. We then computed the differential expression between samples of hSAT and hVAT using DESeq2 (ref. 46). Last, we applied enrichment analyses to the differentially expressed genes of each depot as described above, using clusterProfiler with Kyoto Encyclopedia of Genes and Genomes (KEGG) and GO as reference gene sets (Supplementary Data 2). GSEA was applied to genes ordered by their fold-change values. Enrichment was applied to genes with twofold-change or more and Padj ≤ 0.05 relative to a background set of all genes expressed in adipocytes of that depot.
Comparison between in-house and other hSAT and hVAT adipocyte datasets
We used multiple analyses to test for the existence of nonclassical adipocytes in other datasets. First, we integrated the in-house data with the dataset of Emont et al.9. Specifically, adipocytes were extracted from Emont et al.9 and combined with adipocytes detected in the present study per depot. The combined dataset was reanalyzed, including normalization, determination of variable genes, scaling and integration using Harmony40, with the grouping variable set to tissue source (in-house or Emont et al.) and sample name. Next, to avoid over-clustering, clusters were tested using AssessNodes function from Seurat v.2.0 (thresholds P = 0.08 for hSAT and 0.09 for hVAT). Integrated clusters were annotated based on the dominant adipocyte subtype contributing most of its cells to the cluster. A riverplot analysis matching the Emont et al.9 adipocytes to in-house adipocytes was performed using FindTransferAnchors function from Seurat (with reduction parameter=rpca). Second, we used machine learning to test for the presence of adipocyte subtypes in the Emont et al. dataset9. Per depot, we trained a separate random forest classifier per in-house adipocyte subtype with >200 nuclei. Training was based on the top 20 positive differentially expressed genes for each adipocyte subtype that were also included in the Emont et al. dataset9. Training was assessed using fivefold crossvalidation on 80% of the data and then tested on the remaining data. The area under the receiver operating characteristic curve (auROC) and area under the recall precision curve (auPRC) of each model appear in Supplementary Fig. 9a. Next, we applied each trained model to classify adipocytes of the corresponding depot from Emont et al.9. Adipocytes were annotated to the subtype with the highest probability score >0.2. The prediction score of each model per depot appears in Supplementary Table 6. Third, we applied GSEA to the adipocyte clusters detected by Emont et al.9 and visualized enrichments as described above. Last, we tested whether nonclassical adipocytes could be observed in bulk RNA-seq data of hSAT and hVAT from 73 donors (described below) and in the hSAT spatial transcriptomics dataset of Backdahl et al.11. These analyses are described in Supplementary Note and Supplementary Figs. 9c and 10.
Deconvolution analysis
Analysis was performed using the sNucConv tool16. We trained a separate model for hSAT and hVAT, based on five hSAT and six hVAT samples for which both snRNA-seq and bulk RNA-seq data were available16. Per depot, we trained two separate models, one model treating adipocytes as a single-cell type (used in, for example, Fig. 1d) and the other distinguishing between classical and nonclassical adipocytes (used in, for example, Extended Data Fig. 4). The performance of trained models was assessed using leave one out, by comparing the estimated proportions and true snRNA-seq-derived proportions of the different cell types in the left-out sample. Pearson’s r coefficient was used to quantify the correlation accuracy per model. Trained models obtained r ≥ 0.88 in hSAT and r ≥ 0.93 in hVAT. Trained models were applied using the sNucConv tool to 73 paired bulk RNA-seq profiles of hSAT and hVAT. Specifically, paired omental visceral and abdominal subcutaneous samples were obtained from 73 donors who were selected from the LOBB. Donors included in the deconvolution analysis had a preoperative BMI of 54.5 ± 9.3 kg m−2 and age of 44.1 ± 9.2 years. Type 2 diabetes was diagnosed in 28 patients. For each donor, adipose tissue was collected during elective laparoscopic abdominal surgery, and anthropometric as well as laboratory measurements were obtained, as previously described47. We computed Spearman’s correlations between estimated percentages of cell types per depot (Supplementary Data 6a) and clinical parameters, for parameters with seven or more values (Supplementary Data 6b), with adipocytes split into classical and nonclassical (the latter as a single set). Correlation values appear in Supplementary Data 6c,d.
Microscopic analysis of nonclassical adipocytes
Sections of 5 µm of hSAT and hVAT were deparaffinized, subjected to antigen retrieval (heating in a water bath, 95 °C, 40 min, in antigen retrieval buffer, Abcam, cat. no. ab93680), blocked with 5% BSA for 30 min at room temperature, followed by incubation with primary antibodies overnight, 4 °C (Supplementary Table 7). After washing, sections were incubated with the corresponding secondary antibody (Supplementary Table 7) and nuclei counter stain (DAPI). Microscopy included fluorescence confocal CellDiscoverer 7 (Zeiss) for panoramic scans (×20 objective), and LSM880 confocal microscope (Zeiss) using ×63 oil objective for fluorescent spectrum measurements and imaging single adipocytes. Acquisition parameter thresholds were determined based on omitting primary antibody (negative control) and staining sections of human tissues known to express the respective proteins (positive control), as indicated in the figure legends. Fluorescence spectrum measurements were performed for each primary–secondary antibody pair, to ensure that the signal obtained in adipose tissue sections was identical to that obtained from the positive control (Extended Data Fig. 2).
Trajectory analysis
Trajectory analysis was conducted using Palantir21 v.1.2.0, with the parameters set as in the Palantir tutorial, and using diffusion pseudotime (DPT)22 as implemented in Scanpy48 v.1.9.3 (dpt function). Per analysis, the nuclei corresponding to the relevant cell types were extracted and reanalyzed, including normalization, log transformation, detection of highly variable genes, dimensionality reduction and integration using Harmony. Analyses were based on the 1,500 most variable genes, as suggested by Palantir, except for the analysis of VA and ASPC, where the number of most variable genes was raised to 2,500, to obtain output. The visualization of trajectories was done using ForceAtlas2 (ref. 49). The start cell for the main trajectory analysis in both hSAT and hVAT was an ASPC1 nucleus randomly selected from the 200 cells with the highest DPP4 expression. Trajectory analysis was also applied to the double-filtered dataset of each tissue and using additional ASPC subsets and markers as starting points, which led to similar results (Supplementary Note and Supplementary Figs. 13 and 14).
Intercellular communication analysis
Prediction of enriched intercellular interactions was performed using CellphoneDB27 v.4.0.0 with default parameters. The CellphoneDB heatmap was obtained using the ktplotspy library. For visualization, we summed the ligand–receptor interactions of each pair of cell types (that is, VA2 to VA4 and VA4 to VA2) and plotted it using the Seaborn library. The datasets created by CellphoneDB appear in Supplementary Data 5. Prediction of intercellular communication probabilities and key signaling pathways were performed using CellChat28,50 v.1.6.0 with default parameters, except population.size, which was set to True to minimize the effects of larger cell groups in the data. Analyses were performed per depot, using either adipocytes alone or adipocytes and adipose immune cells. Key signaling pathways were detected using the netAnalysis_signalingRole_network function. A visual representation of the inferred communication network was obtained using the netVisual_aggregate function with chord layout and netVisual_heatmap for heatmap representation. Significant communication interactions and key signaling pathways appear in Supplementary Data 5.
Statistics and reproducibility
Samples were collected from a relatively healthy cohort of men (20%) and women (80%) undergoing elective abdominal surgeries. No statistical method was used to predetermine sample size. Adjustment of P values for multiple hypothesis testing was done using the Benjamini–Hochberg procedure.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Raw snRNA-seq data are available in the Gene Expression Omnibus (GEO), accession no. GSE281356. Processed snRNA-seq data are available in the CZ CELLxGENE portal: https://cellxgene.cziscience.com/collections/ba84c7ba-8d8c-4720-a76e-3ee37dc89f0b. The snRNA-seq dataset of Emont et al.9 was obtained from GEO, accession no. GSE176171. The spatial transcriptomics dataset of Backdahl et al.11 was obtained from https://data.mendeley.com/datasets/3bs5f8mvbs/1. Data of nuc-MT genes were obtained from MitoCarta v.3.0 (ref. 42). Source data are provided with this paper.
Code availability
The code for running the analyses and recreating figures is available via Zenodo at https://doi.org/10.5281/zenodo.14001209 (ref. 51) and https://github.com/Yeger-Lotem-Lab-Code/HCA_Adipocytes_snRNA/tree/v1.0.0.
References
Dressler, W. W. Lifestyle, stress, and blood pressure in a southern black community. Psychosom. Med. 52, 182–198 (1990).
Golan, R. et al. Abdominal superficial subcutaneous fat: a putative distinct protective fat subdepot in type 2 diabetes. Diabetes Care 35, 640–647 (2012).
Ghaben, A. L. & Scherer, P. E. Adipogenesis and metabolic health. Nat. Rev. Mol. Cell Biol. 20, 242–258 (2019).
Lee, M. J., Wu, Y. & Fried, S. K. Adipose tissue heterogeneity: implication of depot differences in adipose tissue for obesity complications. Mol. Asp. Med. 34, 1–11 (2013).
Fain, J. N., Madan, A. K., Hiler, M. L., Cheema, P. & Bahouth, S. W. Comparison of the release of adipokines by adipose tissue, adipose tissue matrix, and adipocytes from visceral and subcutaneous abdominal adipose tissues of obese humans. Endocrinology 145, 2273–2282 (2004).
Ibrahim, M. M. Subcutaneous and visceral adipose tissue: structural and functional differences. Obes. Rev. 11, 11–18 (2010).
Luong, Q., Huang, J. & Lee, K. Y. Deciphering white adipose tissue heterogeneity. Biology (Basel) 8, 23 (2019).
Gesta, S. et al. Evidence for a role of developmental genes in the origin of obesity and body fat distribution. Proc. Natl Acad. Sci. USA 103, 6676–6681 (2006).
Emont, M. P. et al. A single-cell atlas of human and mouse white adipose tissue. Nature 603, 926–933 (2022).
Massier, L. et al. An integrated single cell and spatial transcriptomic map of human white adipose tissue. Nat. Commun. 14, 1438 (2023).
Backdahl, J. et al. Spatial mapping reveals human adipocyte subpopulations with distinct sensitivities to insulin. Cell Metab. 33, 1869–1882 e6 (2021).
Palani, N. P. et al. Adipogenic and SWAT cells separate from a common progenitor in human brown and white adipose depots. Nat. Metab. 5, 996–1013 (2023).
Sun, W. et al. snRNA-seq reveals a subpopulation of adipocytes that regulates thermogenesis. Nature 587, 98–102 (2020).
Sarvari, A. K. et al. Plasticity of epididymal adipose tissue in response to diet-induced obesity at single-nucleus resolution. Cell Metab. 33, 437–453.e5 (2021).
Vijay, J. et al. Single-cell analysis of human adipose tissue identifies depot and disease specific cell types. Nat. Metab. 2, 97–109 (2020).
Sorek, G. et al. sNucConv: a bulk RNA-seq deconvolution method trained on single-nucleus RNA-seq data to estimate cell-type composition of human subcutaneous and visceral adipose tissues. iScience 27, 110368 (2024).
Aibar, S. et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086 (2017).
Toyosaki, M. et al. Dermal fibroblast-like cells reprogrammed directly from adipocytes in mouse. Sci. Rep. 10, 21467 (2020).
Dankel, S. N. et al. COL6A3 expression in adipose tissue cells is associated with levels of the homeobox transcription factor PRRX1. Sci. Rep. 10, 20164 (2020).
Kraunsoe, R. et al. Mitochondrial respiration in subcutaneous and visceral adipose tissue from patients with morbid obesity. J. Physiol. 588, 2023–2032 (2010).
Setty, M. et al. Characterization of cell fate probabilities in single-cell data with Palantir. Nat. Biotechnol. 37, 451–460 (2019).
Haghverdi, L., Buttner, M., Wolf, F. A., Buettner, F. & Theis, F. J. Diffusion pseudotime robustly reconstructs lineage branching. Nat. Methods 13, 845–848 (2016).
Ferrero, R., Rainer, P. & Deplancke, B. Toward a consensus view of mammalian adipocyte stem and progenitor cell heterogeneity. Trends Cell Biol. 30, 937–950 (2020).
Marcelin, G. et al. A PDGFRα-mediated switch toward CD9high adipocyte progenitors controls obesity-Induced adipose tissue fibrosis. Cell Metab. 25, 673–685 (2017).
Deng, T. et al. Class II major histocompatibility complex plays an essential role in obesity-induced adipose inflammation. Cell Metab. 17, 411–422 (2013).
Hagberg, C. E. et al. Flow cytometry of mouse and human adipocytes for the analysis of browning and cellular heterogeneity. Cell Rep. 24, 2746–2756.e5 (2018).
Garcia-Alonso, L. et al. Single-cell roadmap of human gonadal development. Nature 607, 540–547 (2022).
Jin, S. et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 12, 1088 (2021).
Lonnqvist, F., Arner, P., Nordfors, L. & Schalling, M. Overexpression of the obese (ob) gene in adipose tissue of human obese subjects. Nat. Med. 1, 950–953 (1995).
van Harmelen, V. et al. Increased lipolysis and decreased leptin production by human omental as compared with subcutaneous preadipocytes. Diabetes 51, 2029–2036 (2002).
Chun, S. Y. et al. Preparation and characterization of human adipose tissue-derived extracellular matrix growth factors and stem cells: A concise review. Tissue Eng Regen Med. 16, 385–393 (2019).
Hepler, C. et al. Identification of functionally distinct fibro-inflammatory and adipogenic stromal subpopulations in visceral adipose tissue of adult mice. eLife 7, e39636 (2018).
Yu, X. et al. Involution of brown adipose tissue through a Syntaxin 4 dependent pyroptosis pathway. Nat. Commun. 15, 2856 (2024).
Haim, Y. et al. Elevated autophagy gene expression in adipose tissue of obese humans: a potential non-cell-cycle-dependent function of E2F1. Autophagy 11, 2074–2088 (2015).
Maixner, N. et al. A TRAIL-TL1A paracrine network involving adipocytes, macrophages, and lymphocytes induces adipose tissue dysfunction downstream of E2F1 in human obesity. Diabetes 69, 2310–2323 (2020).
Fleming, S.J. et al. Unsupervised removal of systematic background noise from droplet-based single-cell experiments using CellBender. Nat. Methods 20, 1323–1335 (2023).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019).
McGinnis, C. S., Murrow, L. M. & Gartner, Z. J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 8, 329–337.e4 (2019).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Xu, C. et al. Automatic cell type harmonization and integration across Human Cell Atlas datasets. Cell 186, 5876–5891 (2023).
Rath, S. et al. MitoCarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 49, D1541–D1547 (2021).
Wu, T. et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (Camb.) 2, 100141 (2021).
Eden, E., Navon, R., Steinfeld, I., Lipson, D. & Yakhini, Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinf. 10, 48 (2009).
Bhaduri, A., Nowakowski, T. J., Pollen, A. A. & Kriegstein, A. R. Identification of cell types in a mouse brain single-cell atlas using low sampling coverage. BMC Biol. 16, 113 (2018).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Kloting, N. et al. Insulin-sensitive obesity. Am. J. Physiol. Endocrinol. Metab. 299, E506–E515 (2010).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Jacomy, M., Venturini, T., Heymann, S. & Bastian, M. ForceAtlas2, a continuous graph layout algorithm for handy network visualization designed for the Gephi software. PLoS ONE 9, e98679 (2014).
Wolock, S. L., Lopez, R. & Klein, A. M. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 8, 281–291.e9 (2019).
Lazarescu, O. et al. Yeger-Lotem-Lab-Code/HCA Adipocytes snRNA-sequencing atlas. Zenodo https://doi.org/10.5281/zenodo.14001209 (2024).
Acknowledgements
This publication is part of the Human Cell Atlas: www.humancellatlas.org/publications. The present study has been made possible in part by grant nos. 2019-002441 and 2021-239071 from the Chan Zuckerberg Initiative Foundation. This work was also funded by the Deutsche Forschungsgemeinschaft (German Research Foundation) through CRC 1052, project no. 209933838, subproject B1 (to M.B.), B2 (to A.R.), B9 (to M.G.) and C5 (to A.K.). N.H. holds the Goren-Khazzam Chair in Neurobiology and is supported by the Myers Foundation. We thank N. Ofir and T. Ulanovski for their technical help in the cluster preservation analysis. We thank N. Izchaki and P. A. Nono Nankam for their valuable input.
Author information
Authors and Affiliations
Contributions
I.H., A.K., R.M., M.B., N.H., A.R. and E.Y.-L. conceived the project. O.L., M.Z.-A., Y.H., J.J., D.K., L.L., M.G., N.H., A.R. and E.Y.-L. performed the methodology. O.L., M.Z.-A., Y.H., A.S., H.M., A.Z., M.R.-L., L.L., M.A., J.B., C.M., N.R. and M.G. did the investigations. O.L., M.Z.-A., Y.H., A.S., A.R. and E.Y.-L. wrote the original draft of the manuscript. O.L., M.Z.-A., Y.H., A.S., M.G., A.K., R.M., M.B., N.H., A.R. and E.Y.-L. reviewed and edited the manuscript. A.K., R.M., M.B., N.H., A.R. and E.Y.-L. acquired the funding. I.F.L., U.Y., O.D., A.K., R.M., M.B. and A.R. provided resources. A.R. and E.Y.-L. supervised the project.
Corresponding authors
Ethics declarations
Competing interests
M.B. received honoraria as a consultant and speaker from Amgen, AstraZeneca, Bayer, Boehringer-Ingelheim, Daiich-Sankyo, Lilly, Novo Nordisk, Novartis, Pfizer and Sanofi. All other authors declare no competing interests.
Peer review
Peer review information
Nature Genetics thanks Shalev Itzkovitz, Kosaku Shinoda and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 UMAPs of the different markers used to identify adipocyte subpopulations in adipose tissue.
A-B. UMAPs of hSAT (A) and hVAT (B), colored by cell type. C-D. UMAPs of hSAT (C) and hVAT (D), colored by the expression of different markers used for the identification of the adipocyte subpopulations. Red arrows indicate, when apparent, zones in the adipocyte cluster expressing the specified gene. (The expression of these markers in UMAPs of adipocyte clusters is presented in Fig. 5).
Extended Data Fig. 2 Ensuring specificity of fluorescent signal in immunofluorescence analyses of non-classical adipocytes.
A-E. Representative images showing tissues expressing the specific marker used as a positive control (PTPRB – stomach (A); PDE4D – lymph node (B); SKAP1 – lymph node (C); ANK2 – kidney (D); MT-CO2 - colon (C)), no-primary antibody as a negative control, and the positive signal from within stained adipose tissue. The fluorescence emission spectra obtained from these slides are displayed in the graphs. White squares indicate the areas where spectrum measurements were taken. Shown are results of a single experiment per antibody. F. Images highlighting the presence of crown-like structures (CLS) within adipose tissue, visualized using immunofluorescence staining for PLIN1 (green), PDE4D (red) and DAPI (blue). CLS in adipose tissue act as an intrinsic positive control for PDE4D staining, displaying weak signal for PLIN1, accompanied by robust DAPI staining.
Extended Data Fig. 3 Examples of non-classical adipocytes from additional patients.
A-C. Representative images of adipocytes displaying positive signal for the different markers used to identify the different adipocytes subpopulations (PDE4D (A); SKAP1 (B); ANK2 (C)) taken from additional patients, with an additional zoomed out images to illustrate prevalence and localization. Thin scale bar = 20 μm. Thick scale bar = 200 μm. D. Similar analyses for validating VA6 using the protein marker encoded by the mitochondrial gene MT-CO2, with CD36, instead of PLIN1, for staining adipocytes (*), for technical (antibody compatibility) purposes. Shown are representative images of adipocytes from 6-8 different patients (from at least 2 independent staining experiments).
Extended Data Fig. 4 Depot-specific correlations between estimated percentages of classical or non-classical adipocytes and clinical parameters.
Percentages of classical and non-classical adipocytes in each depot separately were estimated in an independent set of samples analyzed by bulk RNA-seq (n = 73), using sNucConv deconvolution algorithm (a validated deconvolution tool for human adipose tissue based on paired samples sequenced by both bulk and snRNA-seq – see Methods for more detail). The percentage of classical adipocytes in hSAT was negatively correlated with waist-to-hip ratio (WHR) and HOMA-IR (Spearman r = -0.4 and -0.55, adjusted p = 0.17 and 0.06, respectively). The percentage of non-classical adipocytes in hSAT was positively correlated with WHR and HOMA-IR (Spearman r = 0.34 and 0.6, adjusted p = 0.17 and 0.05, respectively). The percentage of classical adipocytes in hVAT was negatively correlated with decease in triglycerides (TG) post bariatric surgery (BS) (Spearman r = 0.49, adjusted p = 0.1). Shaded grey areas mark a confidence interval of 0.95. P-values were adjusted using Benjamini-Hochberg procedure.
Extended Data Fig. 5 CD45 (PTPRC) identification as a marker for SA/VA 3 + 4 positive adipocytes in human adipose tissue using different approaches.
A. Representative IHC images of hSAT and hVAT sections stained with anti-CD45 Ab, with and without counterstaining to highlight the positive signal from the adipocytes. scale bar = 20 μm. Shown are representative images of adipocytes from 6-8 different patients (from at least 2 independent staining experiments). B. panel of representative images showing lymph node section expressing the specific marker used as a positive control, no-primary antibody as a negative control, and the positive signal from within stained adipose tissue. The fluorescence emission spectra obtained from these slides are displayed in the graph. White squares indicate the areas where spectrum measurements were taken. Shown are results of a single experiment. C-D. Representative images of adipocytes displaying positive signal for CD45 taken from hSAT and hVAT of two different patients (each panel shows a different patient). scale bar = 20 μm. These 2 patients are representative images of adipocytes from 6-8 different patients (from at least 2 independent staining experiments). E. Image-stream flow cytometry analyses of isolated human adipose tissue cells. PTPRC protein (CD45, common leukocyte antigen) is shown in SVF cells, potentially macrophages, and CD45-negative adipocytes (based on round morphology, diffuse Bodipy neutral lipids staining, CD36-postive, nuclear Draq5-negative6, and with typically polarized light scattering). Shown are results of 1 of 2 independent experiments.
Extended Data Fig. 6 Expression of adiponectin, leptin, and their receptors in hSAT and hVAT.
A. Expression levels in adipocyte subtypes of hSAT (left) and hVAT (right). B. Expression levels in non-adipocyte cell-types of each depot.
Extended Data Fig. 7 Mast cells involvement as source for ECM, fibrotic and inflammatory pathways differs between depots.
A. In hVAT but not in hSAT, mast cells are senders of ECM and fibrotic pathways SEMA3, COLLAGEN and LAMININ. B. In hVAT but not in hSAT, mast cell are senders of the pro-inflammatory (IL16) signaling pathway.
Supplementary information
Supplementary Information
Supplementary Note and Figs. 1–17.
Supplementary Tables
Supplementary Tables 1–7.
Supplementary Data 1
Supplementary data.
Supplementary Data 2
Supplementary data.
Supplementary Data 3
Supplementary data.
Supplementary Data 4
Supplementary data.
Supplementary Data 5
Supplementary data.
Supplementary Data 6
Supplementary data.
Source Data for Supplementary Figs.
Source data for Supplementary file.
Source data
Source Data Fig. 1
Source data for Fig. 1.
Source Data Extended Data Fig. 4
Source data for Extended Data Fig. 4.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lazarescu, O., Ziv-Agam, M., Haim, Y. et al. Human subcutaneous and visceral adipocyte atlases uncover classical and nonclassical adipocytes and depot-specific patterns. Nat Genet 57, 413–426 (2025). https://doi.org/10.1038/s41588-024-02048-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-024-02048-3
This article is cited by
-
Towards a consensus atlas of human and mouse adipose tissue at single-cell resolution
Nature Metabolism (2025)