Study reveals how transcription factors navigate DNA architecture to shape cellular identity

A new study led by Prof. Yosef Buganim from the Hebrew University of Jerusalem and Dr. Abdenour Soufi from the University of Edinburgh reveals how transcription factors (TFs)—key proteins that regulate gene activity—navigate DNA and chromatin structures to determine cellular identity. This discovery provides new insights into how cells establish their roles and opens pathways for advancements in regenerative medicine and cell therapy.
Transcription factors are proteins that bind to specific DNA sequences to control gene expression, guiding cells to become specific types—such as skin, muscle, or placenta cells. While TFs are known to recognize DNA sequences, the process by which they identify their precise targets across the vast genome remained unclear. This study uncovers "guided search" mechanisms, where the 3D architecture of DNA and chromatin—which packages and organizes genetic material—acts as a signpost to direct TFs to the correct genes.
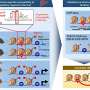
Using cutting-edge technologies, the researchers examined how combinations of TFs drive distinct cell identities, such as embryo versus placenta cells. Their results revealed that transcription factors dynamically cooperate or compete, depending on the chromatin landscape, to precisely target genes critical for defining cell type.
A significant discovery was the role of chromatin topology—the folding and looping of DNA within the nucleus. TFs were shown to follow DNA patterns and chromatin loops as pathways to locate target genes or cluster at key chromatin junctions tightly packed with DNA motifs. Novel DNA elements were identified as genomic signposts that guide TFs to the correct genetic switches necessary for activating cell-type-specific genes.
The study introduces new "guided search" models, demonstrating how the spatial arrangement of chromatin directs different TFs to their relevant targets, offering a deeper understanding of how cellular identity is formed and maintained.
Prof. Buganim stated, "By uncovering how transcription factors interact with chromatin architecture, we can better understand gene regulation and cellular identity. This knowledge opens exciting possibilities for regenerative medicine, enabling us to precisely control cell fate and develop therapies for diseases caused by cellular dysfunction."
The findings pave the way for innovative strategies to manipulate gene expression, with profound implications for regenerative therapies and developmental biology.
The research, titled "Nucleosome fibre topology guides transcription factor binding to enhancers," is published in Nature and provides a powerful framework for exploring gene regulation mechanisms. It holds great promise for understanding age-related diseases, developmental disorders, and advancing the field of cell reprogramming.
More information: Michael R. O'Dwyer et al, Nucleosome fibre topology guides transcription factor binding to enhancers, Nature (2024). DOI: 10.1038/s41586-024-08333-9
Journal information: Nature
Provided by Hebrew University of Jerusalem